7QAW
 
 | |
7QAX
 
 | |
7QAU
 
 | |
7QAQ
 
 | |
7QB8
 
 | |
8RH3
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to Gemcitabine | | Descriptor: | (2~{R},3~{R})-4,4-bis(fluoranyl)-2-(hydroxymethyl)oxolan-3-ol, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
6SD8
 
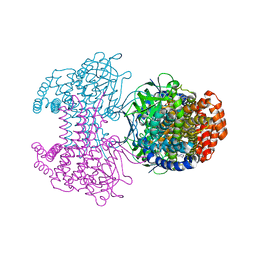 | | Bd2924 apo-form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6SDA
 
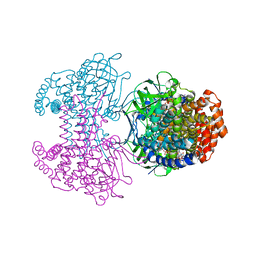 | | Bd2924 C10 acyl-coenzymeA bound form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase, decanoyl-CoA | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
8PZ0
 
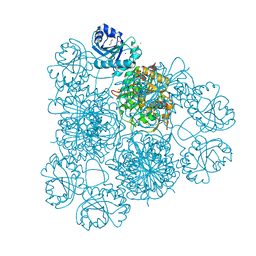 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PZM
 
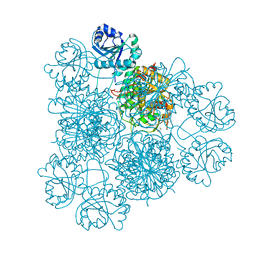 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14 bound to bestatin inhibitor and manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-27 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PQQ
 
 | |
8PZY
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14 - hexameric assembly with manganese bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, AMMONIUM ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-27 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PQR
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to DAD_Immucillin-H | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
8PQT
 
 | |
8PQS
 
 | |
8PQP
 
 | |
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6CP8
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
6CP9
 
 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
