5FIR
 
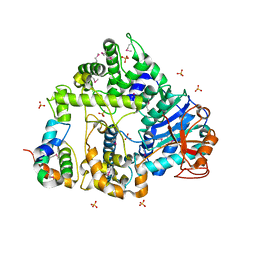 | | Crystal structure of C. elegans XRN2 in complex with the XRN2-binding domain of PAXT-1 | | Descriptor: | 5'-3' EXORIBONUCLEASE 2 HOMOLOG, PAXT-1, SULFATE ION | | Authors: | Richter, H, Katic, I, Gut, H, Grosshans, H. | | Deposit date: | 2015-10-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural Basis and Function of Xrn2-Binding by Xtb Domains
Nat.Struct.Mol.Biol., 23, 2016
|
|
6FQ1
 
 | |
6FQR
 
 | |
6GX6
 
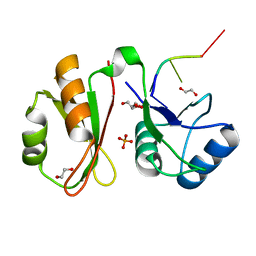 | | Crystal structure of IMP3 RRM12 in complex with RNA (ACAC) | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor 2 mRNA-binding protein 3, PHOSPHATE ION, ... | | Authors: | Jia, M, Gut, H, Chao, A.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IMP3 RRM12 recognition of RNA.
RNA, 24, 2018
|
|
6I3H
 
 | | Crystal structure of influenza A virus M1 N-terminal domain (G18A mutation) | | Descriptor: | Matrix protein 1, PHOSPHATE ION | | Authors: | Miyake, Y, Keusch, J.J, Decamps, L, Ho-Xuan, H, Iketani, S, Gut, H, Kutay, U, Helenius, A, Yamauchi, Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Nat Microbiol, 4, 2019
|
|
5M1X
 
 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
6QSZ
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5G0H
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | Descriptor: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0F
 
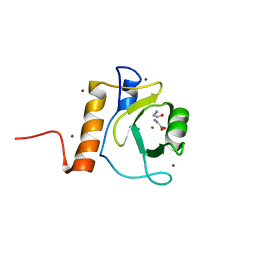 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | Descriptor: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0J
 
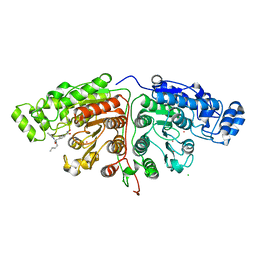 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | Descriptor: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
6RR0
 
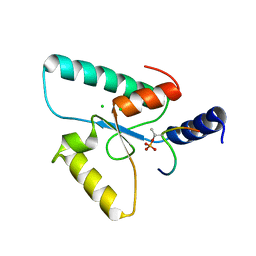 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
6RRV
 
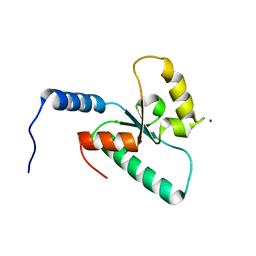 | | Crystal structure of the Sir4 H-BRCT domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Regulatory protein SIR4 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5G0I
 
 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker cleaved) in complex with Nexturastat A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HDAC6, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0G
 
 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | Descriptor: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
4BJT
 
 | | Crystal structure of the Rap1 C-terminal domain (Rap1-RCT) in complex with the Rap1 binding module of Rif1 (Rif1-RBM) | | Descriptor: | 1,2-ETHANEDIOL, DNA-BINDING PROTEIN RAP1, TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ6
 
 | | Crystal structure Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, RAP1-INTERACTING FACTOR 2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ1
 
 | | Crystal structure of Saccharomyces cerevisiae RIF2 | | Descriptor: | CHLORIDE ION, PROTEIN RIF2 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4A0A
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A09
 
 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4AP5
 
 | | Crystal structure of human POFUT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
4AE8
 
 | | Crystal structure of human THEM4 | | Descriptor: | THIOESTERASE SUPERFAMILY MEMBER 4 | | Authors: | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
4BJ5
 
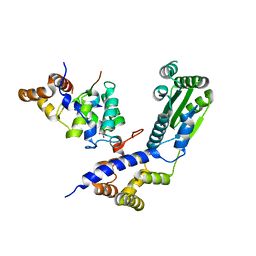 | | Crystal structure of Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, PROTEIN RIF2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4A11
 
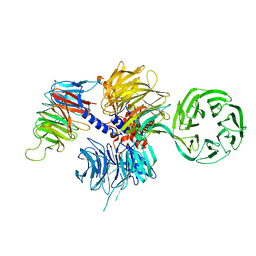 | | Structure of the hsDDB1-hsCSA complex | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | Authors: | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0K
 
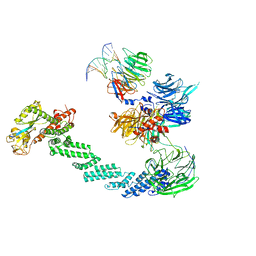 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
