4IMG
 
 | |
4IMC
 
 | |
4IMD
 
 | |
4IMF
 
 | |
2IHJ
 
 | |
2IHZ
 
 | |
2II6
 
 | |
2ILV
 
 | |
2NN3
 
 | | structure of pro-sf-caspase-1 | | Descriptor: | Caspase-1 | | Authors: | Fisher, A.J, Ni, L. | | Deposit date: | 2006-10-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pro-sfcapsase-1, structural insights into activation mechanism of caspases
To be Published
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
4DBC
 
 | | Substrate Activation in Aspartate Aminotransferase | | Descriptor: | (E)-N-{2-hydroxy-3-methyl-6-[(phosphonooxy)methyl]benzylidene}-L-aspartic acid, 1,2-ETHANEDIOL, Aspartate aminotransferase, ... | | Authors: | Toney, M.D, Fisher, A.J, Griswold, W.R. | | Deposit date: | 2012-01-14 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase.
J.Am.Chem.Soc., 134, 2012
|
|
8UUC
 
 | | Crystal structure of a bacterial clusterless MutYX bound to an Abasic site analog (THF) opposite d(8-oxo-G) | | Descriptor: | 1,2-ETHANEDIOL, Adenine DNA glycosylase, CHLORIDE ION, ... | | Authors: | Trasvina-Arenas, C.H, David, S.S, Fisher, A.J. | | Deposit date: | 2023-11-01 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of MutYX: A novel clusterless adenine DNA glycosylase with a distinct C-terminal domain and 8-Oxoguanine recognition sphere.
Biorxiv, 2025
|
|
6BN0
 
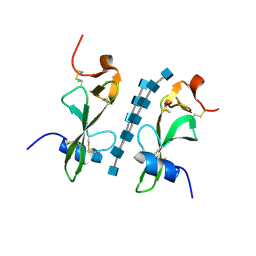 | | Avirulence protein 4 (Avr4) from Cladosporium fulvum bound to the hexasaccharide of chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Race-specific elicitor A4 | | Authors: | Hurlburt, N.K, Fisher, A.J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein.
PLoS Pathog., 14, 2018
|
|
6CKK
 
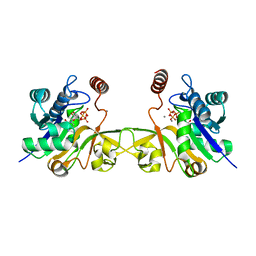 | |
6CKM
 
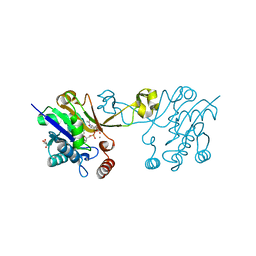 | |
6CKJ
 
 | |
6D06
 
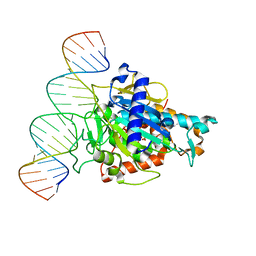 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
6CKL
 
 | | N. meningitidis CMP-sialic acid synthetase in the presence of CMP and Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Matthews, M.M, Fisher, A.J, Chen, X. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Catalytic Cycle ofNeisseria meningitidisCMP-Sialic Acid Synthetase Illustrated by High-Resolution Protein Crystallography.
Biochemistry, 2019
|
|
1I3P
 
 | |
1I3S
 
 | | THE 2.7 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A MUTATED BACULOVIRUS P35 AFTER CASPASE CLEAVAGE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, EARLY 35 KDA PROTEIN | | Authors: | dela Cruz, W.P, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage.
J.Biol.Chem., 276, 2001
|
|
5HP2
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AU basepair at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*UP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HP3
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AC mismatch at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
9BS2
 
 | | Glycosylase MutY variant R149Q in complex with DNA containing d(8-oxo-G) paired with a product analog (THF) to 1.51 A resolution | | Descriptor: | ACETIC ACID, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Trasvina-Arenas, C.H, Tamayo, N, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2024-05-12 | | Release date: | 2024-10-30 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of human MUTYH and functional profiling of cancer-associated variants reveal an allosteric network between its [4Fe-4S] cluster cofactor and active site required for DNA repair.
Nat Commun, 16, 2025
|
|
1I2D
 
 | |
1JHD
 
 | | Crystal Structure of Bacterial ATP Sulfurylase from the Riftia pachyptila Symbiont | | Descriptor: | BROMIDE ION, SULFATE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Beynon, J.D, MacRae, I.J, Huston, S.L, Nelson, D.C, Segel, I.H, Fisher, A.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ATP sulfurylase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila.
Biochemistry, 40, 2001
|
|
