7UZ8
 
 | |
6U5N
 
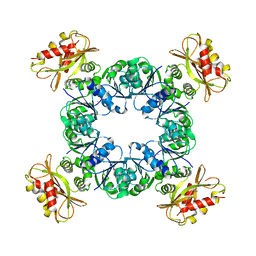 | | Calcium-bound MthK gating ring state 2 | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK | | Authors: | Fan, C, Nimigean, C.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
7UZA
 
 | |
7UZ5
 
 | |
7UZ6
 
 | |
7UZ4
 
 | |
7UZB
 
 | |
7UZ7
 
 | |
7UZC
 
 | |
7UZD
 
 | |
7N5B
 
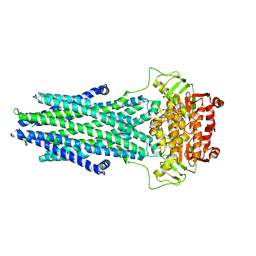 | | Structure of AtAtm3 in the outward-facing conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7N58
 
 | | Structure of AtAtm3 in the inward-facing conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
7N5A
 
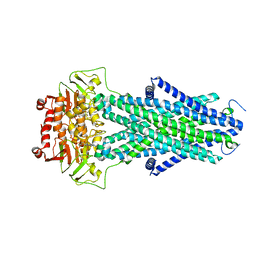 | | Structure of AtAtm3 in the closed conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
4GRU
 
 | |
4GRR
 
 | | characterization of N- and C- terminus mutants of human MIF | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Characterization of N- and C- terminus mutants of human MIF
To be Published
|
|
4GUM
 
 | | Cystal structure of locked-trimer of human MIF | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-07-03 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GRQ
 
 | | Characterization of N- and C- terminus mutants of human MIF | | Descriptor: | 2-methyl-1-[2-(propan-2-yl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-26 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of N- and C- terminus mutants of human MIF
To be Published
|
|
4GRO
 
 | |
4GRP
 
 | |
4GRN
 
 | |
4GUK
 
 | | New crystal form structure of human NCS1 | | Descriptor: | (2S,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YLPHOSPHONI C ACID, 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, ... | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | new crystal form structure of human NCS1
To be Published
|
|
3VA7
 
 | | Crystal structure of the Kluyveromyces lactis Urea Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, GLYCEROL, KLLA0E08119p, ... | | Authors: | Fan, C, Xiang, S. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction
J.Biol.Chem., 287, 2012
|
|
4IST
 
 | |
4ISS
 
 | |
1QQQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SER254 MUTANT OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Fantz, C, Shaw, D, Jennings, W, Forsthoefel, A, Kitchens, M, Phan, J, Minor, W, Lebioda, L, Berger, F.G, Spencer, H.T. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug-resistant variants of Escherichia coli thymidylate synthase: effects of substitutions at Pro-254.
Mol.Pharmacol., 57, 2000
|
|
