4D1Q
 
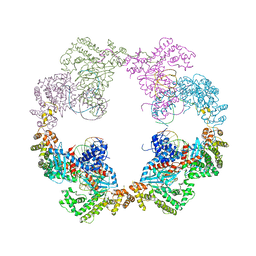 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
7MGV
 
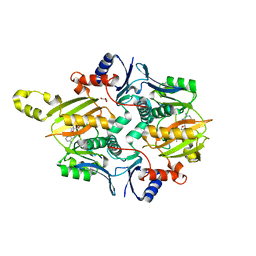 | | Chryseobacterium gregarium RiPP-associated ATP-grasp ligase in complex with ADP, and a leader and core peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CdnA3 Core peptide, CdnA3 Leader peptide, ... | | Authors: | Bewley, C.A, Zhao, G, Kosek, D, Dyda, F. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Basis for a Dual Function ATP Grasp Ligase That Installs Single and Bicyclic omega-Ester Macrocycles in a New Multicore RiPP Natural Product.
J.Am.Chem.Soc., 143, 2021
|
|
7LCC
 
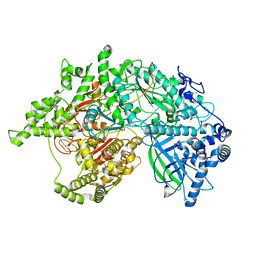 | | Helitron transposase bound to LTS | | Descriptor: | Helraiser K1068Q, LTS, ZINC ION | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2021-01-10 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.
Mol.Cell, 81, 2021
|
|
1T0F
 
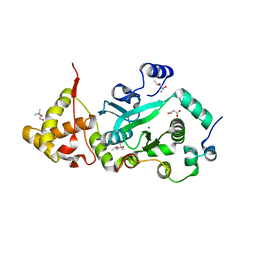 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | Authors: | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | Deposit date: | 2004-04-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
1RZ9
 
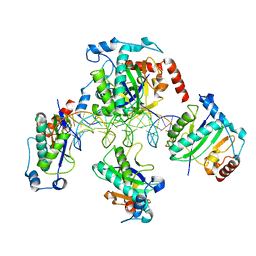 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
3ZYS
 
 | | Human dynamin 1 deltaPRD polymer stabilized with GMPPCP | | Descriptor: | DYNAMIN-1, INTERFERON-INDUCED GTP-BINDING PROTEIN MX1 | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
3ZYC
 
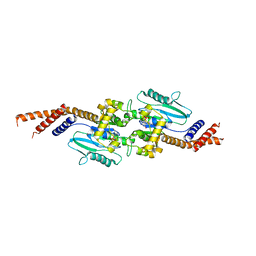 | | DYNAMIN 1 GTPASE GED FUSION DIMER COMPLEXED WITH GMPPCP | | Descriptor: | DYNAMIN-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
1YQV
 
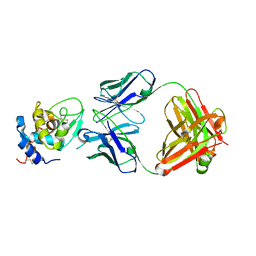 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A6O
 
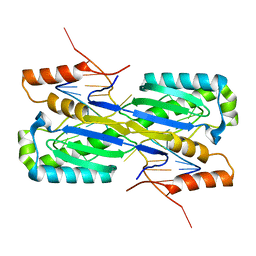 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
2A6M
 
 | | Crystal Structure of the ISHp608 Transposase | | Descriptor: | ISHp608 transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
2XM3
 
 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XO6
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE Y132F MUTANT COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*G)-3', ACETATE ION, CADMIUM ION, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XQC
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XMA
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2BZF
 
 | | Structural basis for DNA bridging by barrier-to-autointegration factor (BAF) | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*GP*AP*GP*GP)-3', BARRIER-TO-AUTOINTEGRATION FACTOR | | Authors: | Bradley, C.M, Ronning, D.R, Ghirlando, R, Craigie, R, Dyda, F. | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for DNA Bridging by Barrier-to-Autointegration Factor.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BW3
 
 | | Three-dimensional structure of the Hermes DNA transposase | | Descriptor: | TRANSPOSASE | | Authors: | Hickman, A.B, Perez, Z.N, Zhou, L, Musingarimi, P, Ghirlando, R, Hinshaw, J.E, Craig, N.L, Dyda, F. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of a Eukaryotic DNA Transposase
Nat.Struct.Mol.Biol., 12, 2005
|
|
1IB1
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3 ZETA:SEROTONIN N-ACETYLTRANSFERASE COMPLEX | | Descriptor: | 14-3-3 ZETA ISOFORM, COA-S-ACETYL TRYPTAMINE, SEROTONIN N-ACETYLTRANSFERASE | | Authors: | Obsil, T, Ghirlando, R, Klein, D.C, Ganguly, S, Dyda, F. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation.
Cell(Cambridge,Mass.), 105, 2001
|
|
1M55
 
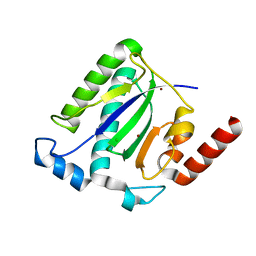 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | Descriptor: | CHLORIDE ION, Rep protein, ZINC ION | | Authors: | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
1MPE
 
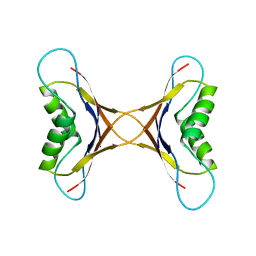 | |
1BIS
 
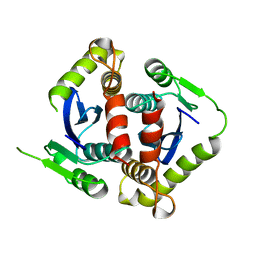 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
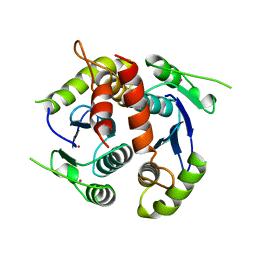 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AER
 
 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH BETA-TAD | | Descriptor: | 2-(1,5-DIDEOXYRIBOSE)-4-AMIDO-THIAZOLE, ADENOSINE MONOPHOSPHATE, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D.R. | | Deposit date: | 1995-12-11 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1BIU
 
 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2VIH
 
 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
