2QM4
 
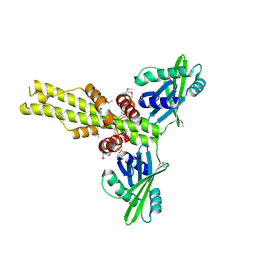 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
2WVI
 
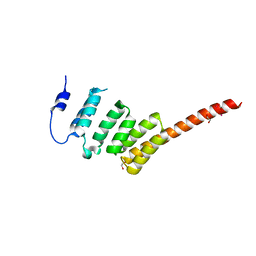 | | Crystal Structure of the N-terminal Domain of BubR1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1 BETA, ... | | Authors: | D'Arcy, S, Davies, O.R, Blundell, T.L, Bolanos-Garcia, V.M. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the Molecular Basis of Bubr1 Kinetochore Interactions and Anaphase-Promoting Complex/Cyclosome (Apc/C)-Cdc20 Inhibition
J.Biol.Chem., 285, 2010
|
|
4CPC
 
 | |
7BGF
 
 | |
8OKH
 
 | |
8PUZ
 
 | |
8PV0
 
 | |
9FF9
 
 | |
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
6SD8
 
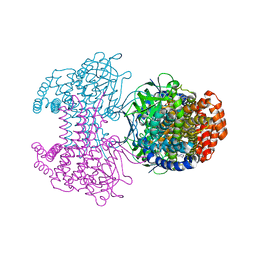 | | Bd2924 apo-form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6SDA
 
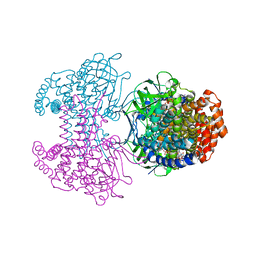 | | Bd2924 C10 acyl-coenzymeA bound form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase, decanoyl-CoA | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6CVZ
 
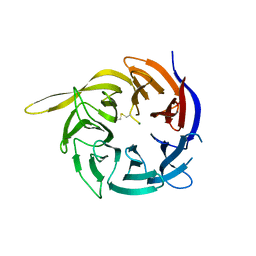 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6CP8
 
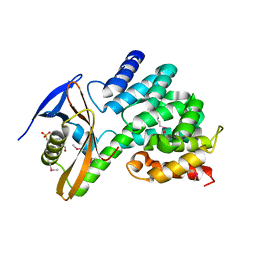 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
6CP9
 
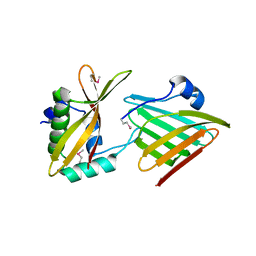 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
