4YEW
 
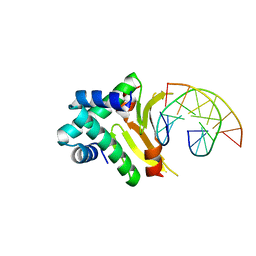 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEY
 
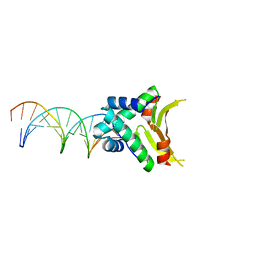 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
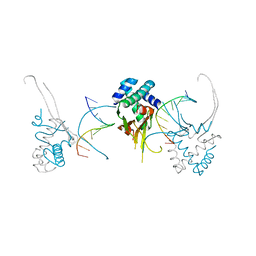 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YF0
 
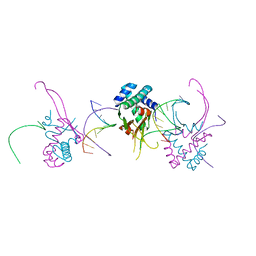 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YDS
 
 | |
4ZBH
 
 | |
4YDD
 
 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
1DWW
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWX
 
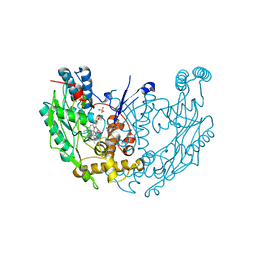 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWV
 
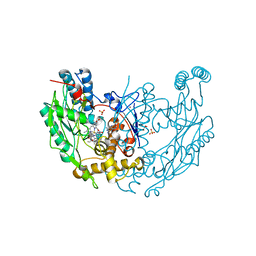 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1UGH
 
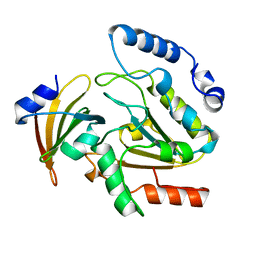 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
3NOD
 
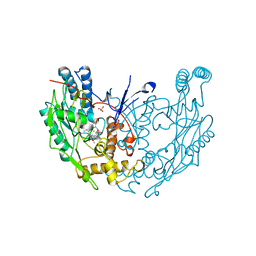 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND PRODUCT ANALOGUE L-THIOCITRULLINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, L-THIOCITRULLINE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
5KSE
 
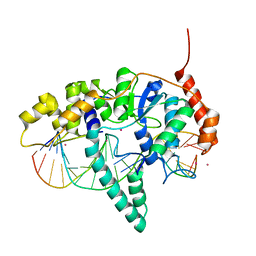 | | Flap endonuclease 1 (FEN1) R100A with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2016-07-08 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
8GJ9
 
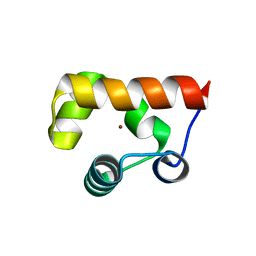 | |
8GJA
 
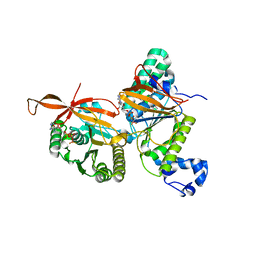 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJ8
 
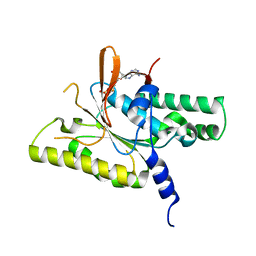 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
6MUJ
 
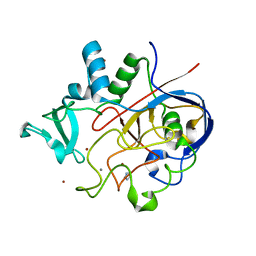 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5K97
 
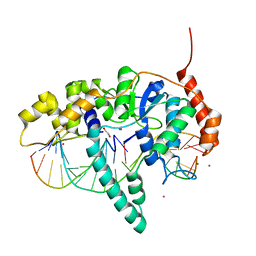 | | Flap endonuclease 1 (FEN1) D233N with cleaved product fragment and Sm3+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
1T38
 
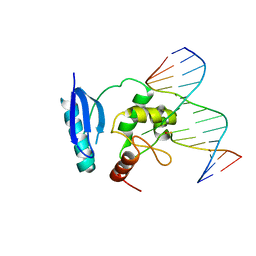 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE BOUND TO DNA CONTAINING O6-METHYLGUANINE | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(6OG)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T39
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE COVALENTLY CROSSLINKED TO DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(E1X)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6X1Z
 
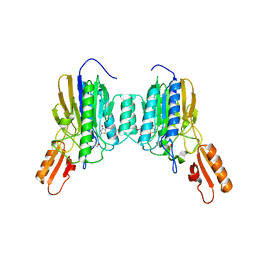 | | Mre11 dimer in complex with small molecule modulator PFMJ | | Descriptor: | (5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6X1Y
 
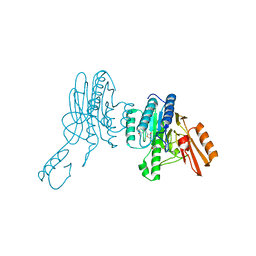 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6WFV
 
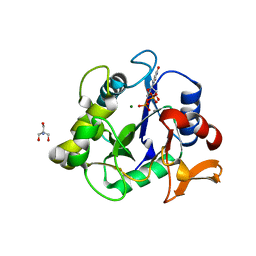 | | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Guo, H.-F, Tsai, C.-L, Miller, M.D, Phillips Jr, G.N, Tainer, J.A, Kurie, J.M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human
To Be Published
|
|
1QOM
 
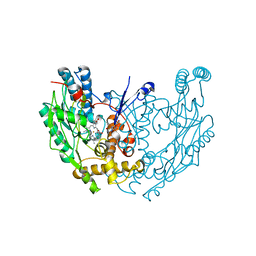 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
5KVH
 
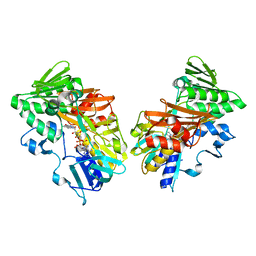 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
