1RCF
 
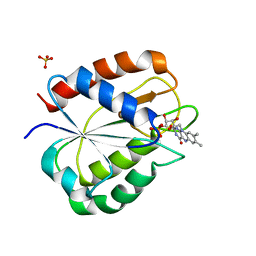 | | STRUCTURE OF THE TRIGONAL FORM OF RECOMBINANT OXIDIZED FLAVODOXIN FROM ANABAENA 7120 AT 1.40 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Burkhart, B, Ramakrishnan, B, Yan, H, Reedstrom, R, Markley, J, Straus, N, Sundaralingam, M. | | Deposit date: | 1994-10-31 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the trigonal form of recombinant oxidized flavodoxin from Anabaena 7120 at 1.40 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1VTF
 
 | |
1OSU
 
 | |
1P79
 
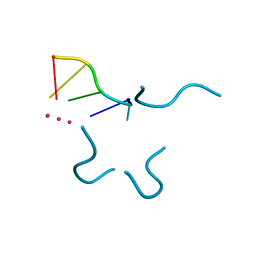 | | Crystal structure of a bulged RNA tetraplex: implications for a novel binding site in RNA tetraplex | | Descriptor: | 5'-R(*UP*GP*UP*GP*GP*U)-3', POTASSIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of a Bulged RNA Tetraplex at 1.1 A Resolution: Implications for a Novel Binding Site in RNA Tetraplex
STRUCTURE, 11, 2003
|
|
217D
 
 | |
216D
 
 | |
213D
 
 | |
246D
 
 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
279D
 
 | |
251D
 
 | |
260D
 
 | |
1R3Z
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*(5CM)P*GP*(5CM)P*GP*CP*GP*C)-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R41
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2AWE
 
 | |
438D
 
 | |
306D
 
 | |
305D
 
 | |
304D
 
 | |
1JZV
 
 | | Crystal structure of a bulged RNA from the SL2 stem-loop of the HIV-1 psi-RNA | | Descriptor: | 5'-R(*CP*AP*GP*UP*AP*CP*GP*(5IC)P*C)-3', 5'-R(*GP*GP*CP*GP*AP*CP*(5BU)P*G)-3', MAGNESIUM ION | | Authors: | Xiong, Y, Sudarsanakumar, C, Deng, J, Pan, B, Sundaralingam, M. | | Deposit date: | 2001-09-17 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Bulged RNA from the SL2 Stem-loop of the HIV-1 psi-RNA
To be Published
|
|
138D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
409D
 
 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
418D
 
 | |
405D
 
 | |
422D
 
 | |
