7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
 | | Nanobody C5 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Weckener, M. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | | Nanobody C5 bound to Kent variant RBD (N501Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5, GLYCEROL, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
6SJ4
 
 | | Amidohydrolase, AHS with substrate analog | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)carbonyloxybenzoic acid, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YQQ
 
 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
6Y47
 
 | |
2VL7
 
 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2VG2
 
 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG3
 
 | | Rv2361 with citronellyl pyrophosphate | | Descriptor: | CHLORIDE ION, GERANYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2V7J
 
 | | PrnB L-tryptophan complex | | Descriptor: | PRNB, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2VG4
 
 | | Rv2361 native | | Descriptor: | UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2V82
 
 | | KDPGal complexed to KDPGal | | Descriptor: | 2-DEHYDRO-3-DEOXY-6-PHOSPHOGALACTONATE ALDOLASE, 2-KETO-DEOXY-GALACTOSE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure of Escherichia coli KDPGal aldolase.
Bioorg. Med. Chem., 16, 2008
|
|
2V7I
 
 | | PrnB native | | Descriptor: | PRNB, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2V7L
 
 | | PrnB 7Cl-L-tryptophan complex | | Descriptor: | 7-CHLOROTRYPTOPHAN, PRNB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily.
Biochemistry, 46, 2007
|
|
2V7K
 
 | | PrnB D-tryptophan complex | | Descriptor: | D-TRYPTOPHAN, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
2V7M
 
 | | PrnB 7-Cl-D-tryptophan complex | | Descriptor: | 7-CL-D-TRYPTOPHAN, PHOSPHATE ION, PRNB, ... | | Authors: | Naismith, J.H. | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway is Related to the Heme-Dependent Dioxygenase Superfamily
Biochemistry, 46, 2007
|
|
7NIH
 
 | | Wzc-K540M MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NHS
 
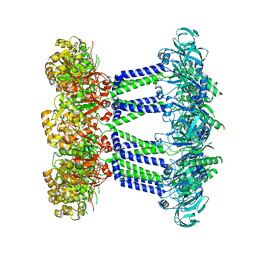 | | Wzc K540M C8 | | Descriptor: | Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NII
 
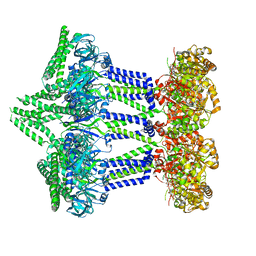 | | Wzc-K540M MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NI2
 
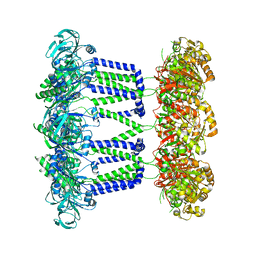 | | Wzc-K540M-4YE C8 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NIB
 
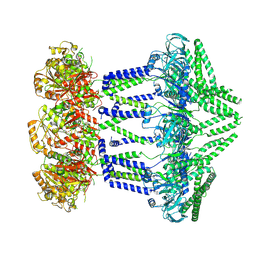 | | Wzc-K540M-4YE C1 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
6FI2
 
 | | VexL: A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid, MALONATE ION, VexL | | Authors: | Naismith, J.H, McMahon, S.A, Le Bas, A, Liston, S.D, Whitfield, C. | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Periplasmic depolymerase provides insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ZHD
 
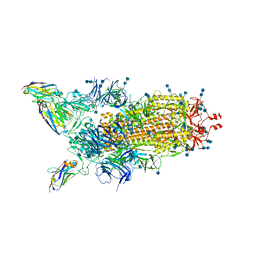 | | H11-H4 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H4, ... | | Authors: | Clare, D.K, Naismith, J.H, Weckener, M, Vogirala, V.K. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | H11-H4 bound to Spike
To Be Published
|
|
6ZH9
 
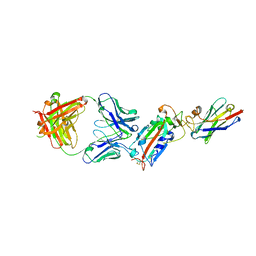 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
