4YZF
 
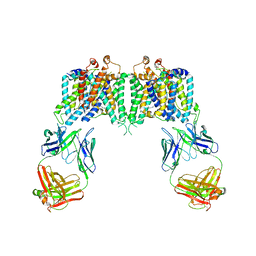 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
5A16
 
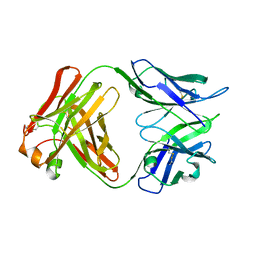 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
5AZD
 
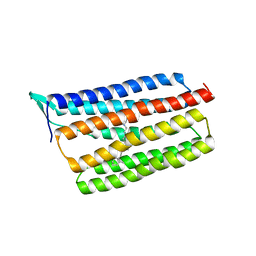 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
5ZE9
 
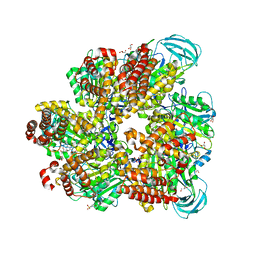 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.384 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKB
 
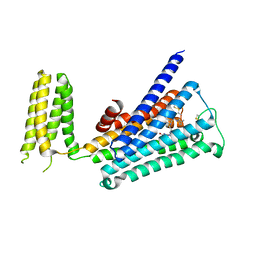 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
3VG9
 
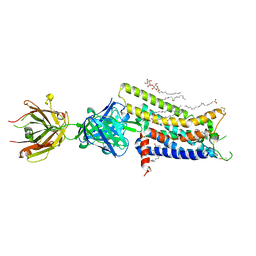 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
3VGA
 
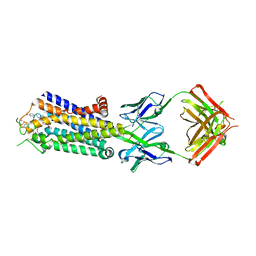 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
7CN1
 
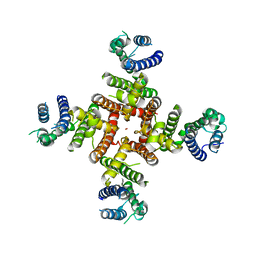 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN0
 
 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7W74
 
 | | Crystal structure of DTG rhodopsin from Pseudomonas putida | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-like protein, ... | | Authors: | Suzuki, K, Konno, M, Bagherzadeh, R, Inoue, K, Murata, T. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural characterization of proton-pumping rhodopsin lacking a cytoplasmic proton donor residue by X-ray crystallography.
J.Biol.Chem., 298, 2022
|
|
7VW7
 
 | | Crystal structure of the 2 ADP-AlF4-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Shekhar, M, Gupta, C, Singharoy, A, Murata, T. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.818 Å) | | Cite: | Revealing a Hidden Intermediate of Rotatory Catalysis with X-ray Crystallography and Molecular Simulations.
Acs Cent.Sci., 8, 2022
|
|
7DNS
 
 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7DHW
 
 | | Crystal structure of myosin-XI motor domain in complex with ADP-ALF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Haraguchi, T, Tamanaha, M, Yoshimura, K, Imi, T, Tominaga, M, Sakayama, H, Nishiyama, T, Ito, K, Murata, T. | | Deposit date: | 2020-11-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of ultrafast myosin, its amino acid sequence, and structural features.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DQE
 
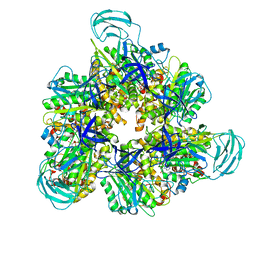 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
3VR2
 
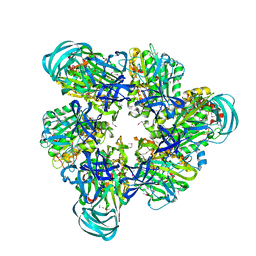 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
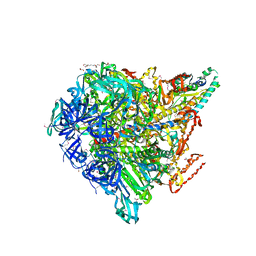 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
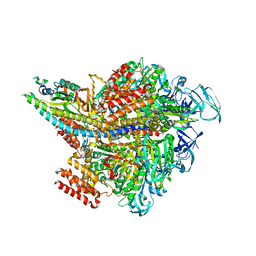 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
