2D58
 
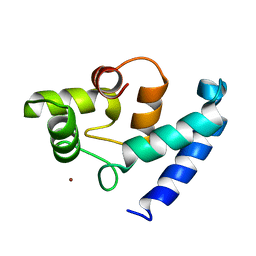 | | Human microglia-specific protein Iba1 | | Descriptor: | Allograft inflammatory factor 1, NICKEL (II) ION | | Authors: | Kamitori, S. | | Deposit date: | 2005-10-31 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
4G09
 
 | |
4G07
 
 | |
5XN2
 
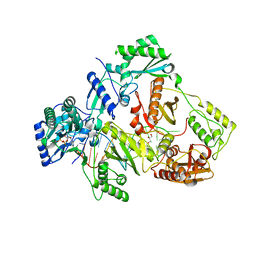 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
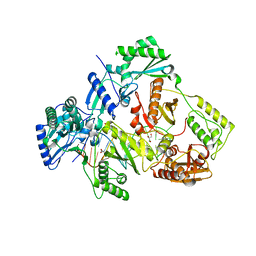 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
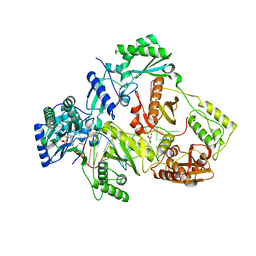 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
6HIR
 
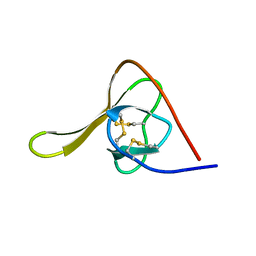 | |
2E3K
 
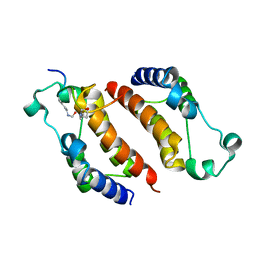 | |
6IK9
 
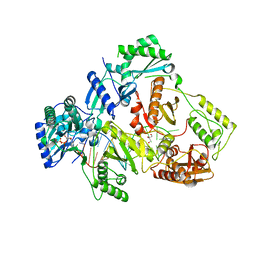 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IKA
 
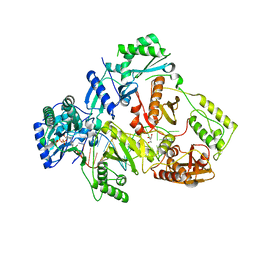 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
4RYD
 
 | |
4HIR
 
 | |
1HIC
 
 | |
2HIR
 
 | |
