2GS9
 
 | | Crystal structure of TT1324 from Thermus thermophilis HB8 | | Descriptor: | FORMIC ACID, Hypothetical protein TT1324, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kamitori, S, Abe, A, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of TT1324 from Thermus thermophilis HB8
To be Published
|
|
8WL7
 
 | |
8WL9
 
 | |
8WL5
 
 | | X-ray structure of Enterobacter cloacae allose-binding protein in free form | | Descriptor: | 1,2-ETHANEDIOL, Allose ABC transporter | | Authors: | Kamitori, S. | | Deposit date: | 2023-09-29 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structures of Enterobacter cloacae allose-binding protein in complexes with monosaccharides demonstrate its unique recognition mechanism for high affinity to allose.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8WLB
 
 | |
3WGP
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (1R,2R,3R,5Z,7E,14beta,17alpha)-2-(3-hydroxypropoxy)-9,10-secocholesta-5,7,10-triene-1,3,25-triol, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Vitamin D receptor
to be published
|
|
1IT9
 
 | | CRYSTAL STRUCTURE OF AN ANTIGEN-BINDING FRAGMENT FROM A HUMANIZED VERSION OF THE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | HUMANIZED ANTIBODY HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Takahashi, T, Miyadai, K, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2002-01-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Humanization of the Mouse Anti-Fas Antibody HFE7A and Crystal Structure of the Humanized HFE7A Fab Fragment
BIOL.PHARM.BULL., 25, 2002
|
|
8YJ8
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, VHH43, ZINC ION | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8YJ5
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, VHH43 | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8YJ6
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, ZINC ION | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8YJ7
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, ZINC ION, scFv13 | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
3AZ1
 
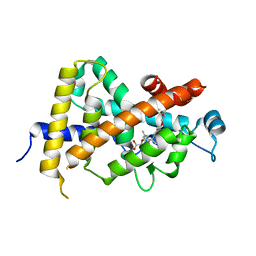 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, {4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ2
 
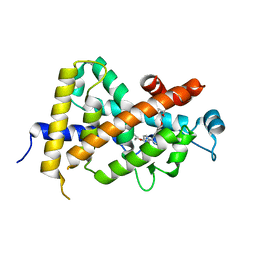 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | 5-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ3
 
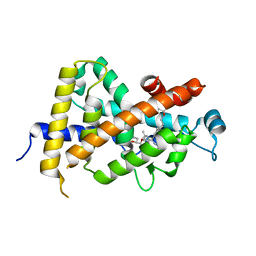 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[4-(3-{4-[(3S)-3-hydroxy-4,4-dimethylpentyl]-3-methylphenyl}pentan-3-yl)-2-methylphenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
4KRU
 
 | |
4KRT
 
 | |
7E2E
 
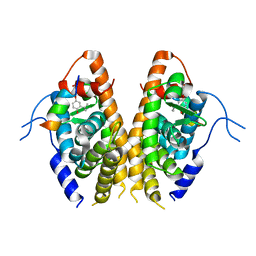 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
7DA2
 
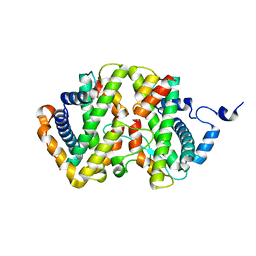 | | The crystal structure of the chicken FANCM-MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Nishino, T, Ito, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analysis of the chicken FANCM-MHF complex and its stability.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2DDQ
 
 | |
3A73
 
 | |
3A5J
 
 | | Crystal structure of protein-tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ito, S. | | Deposit date: | 2009-08-08 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Post-translational modification of a non-catalytic Cys121 of PTP1B
To be Published
|
|
3A5K
 
 | | Crystal structure of protein-tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ito, S. | | Deposit date: | 2009-08-08 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Post-translational modification of a non-catalytic Cys121 of PTP1B
To be Published
|
|
8YXK
 
 | |
8YXN
 
 | |
7F5I
 
 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | Descriptor: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Kamitori, S, Tamai, E. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
