8Q8S
 
 | | Tau - AD-THF | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q8Z
 
 | |
8Q8L
 
 | |
8Q8U
 
 | |
8Q8W
 
 | |
8DM9
 
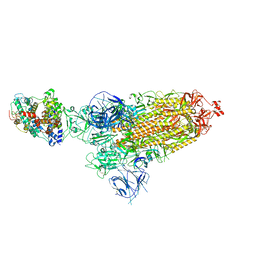 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8Q9A
 
 | |
8Q8E
 
 | |
8Q8Y
 
 | |
8Q9E
 
 | |
8Q8X
 
 | |
8Q9B
 
 | |
8DFS
 
 | | type I-C Cascade bound to AcrIF2 | | Descriptor: | Anti-CRISPR protein 30, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8Q8V
 
 | | Tau - CTE-MIA3 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q9C
 
 | |
8DFO
 
 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DFA
 
 | | type I-C Cascade bound to ssDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8DEX
 
 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8BSD
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Oberthuer, D, Sprenger, J. | | Deposit date: | 2022-11-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
5HIU
 
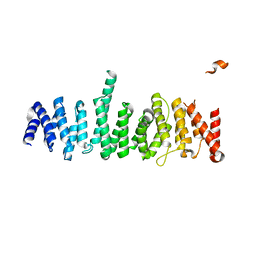 | | Structure of the TSC2 N-terminus | | Descriptor: | GTPase activator-like protein | | Authors: | Zech, R, Kiontke, S, Kummel, D. | | Deposit date: | 2016-01-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Tuberous Sclerosis Complex 2 (TSC2) N Terminus Provides Insight into Complex Assembly and Tuberous Sclerosis Pathogenesis.
J.Biol.Chem., 291, 2016
|
|
6RMQ
 
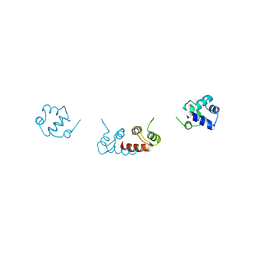 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
3T0Q
 
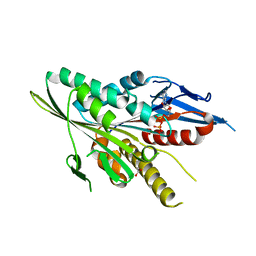 | | Motor Domain Structure of the Kar3-like kinesin from Ashbya gossypii | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, AGR253Wp, ... | | Authors: | Duan, D, Hnatchuk, D.J, Brenner, J, Davis, D, Allingham, J.S. | | Deposit date: | 2011-07-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Kar3-like kinesin motor domain from the filamentous fungus Ashbya gossypii.
Proteins, 80, 2012
|
|
8BZV
 
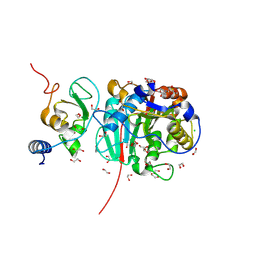 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
3HVJ
 
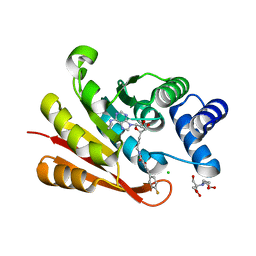 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-propyladenine-containing bisubstrate inhibitor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4BCE
 
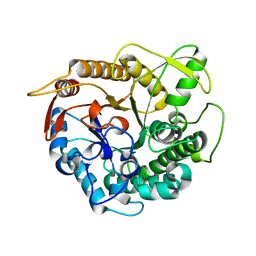 | | crystal structure of Ttb-gly N282T mutant | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Teze, D, Tran, V, Tellier, C, Dion, M, Leroux, C, Roncza, J, Czjzek, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Semi-Rational Approach for Converting a Gh1 Beta-Glycosidase Into a Beta-Transglycosidase.
Protein Eng.Des.Sel., 27, 2014
|
|
