7H5W
 
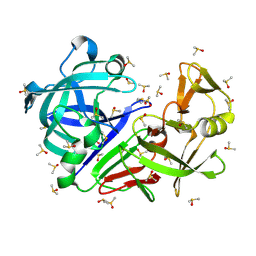 | | Crystal structure of endothiapepsin PF_RT2 in complex with JFD03909 at 296 K | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5N
 
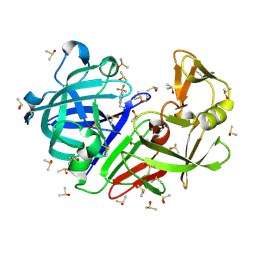 | | Crystal structure of endothiapepsin PN_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5Q
 
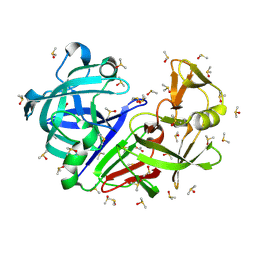 | | Crystal structure of endothiapepsin PF_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5T
 
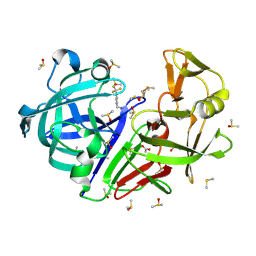 | | Crystal structure of endothiapepsin IS_RT2 in complex with TL00150 at 296 K | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7H5Y
 
 | | Crystal structure of endothiapepsin IS_cryo1 in complex with JFD03909 at 100 K | | Descriptor: | 1,10-PHENANTHROLINE, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Huang, C.-Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-04-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5EX0
 
 | | Crystal structure of human SMYD3 in complex with a MAP3K2 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, MAP3K2 peptide, ... | | Authors: | Fu, W, Liu, N, Qiao, Q, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
5EX3
 
 | | Crystal structure of human SMYD3 in complex with a VEGFR1 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Qiao, Q, Fu, W, Liu, N, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
6KA0
 
 | | Silver-bound E.coli Malate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
5GNB
 
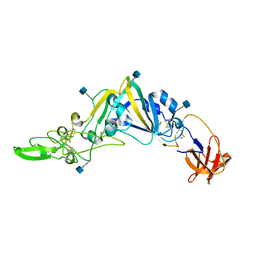 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
6KA1
 
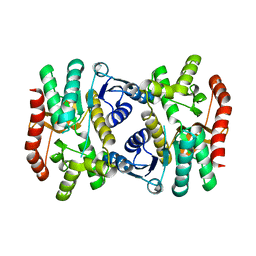 | | E.coli Malate dehydrogenase | | Descriptor: | Malate dehydrogenase | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
7F77
 
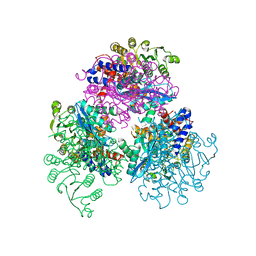 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | Descriptor: | Glutamate dehydrogenase | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F79
 
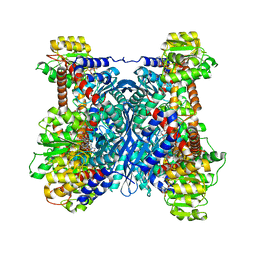 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
5H52
 
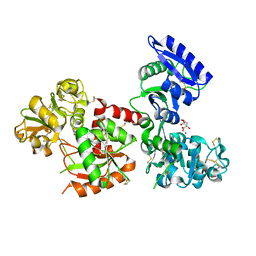 | | Structure of Titanium-bound human serum transferrin | | Descriptor: | CITRIC ACID, MALONATE ION, Serotransferrin, ... | | Authors: | Curtin, J.P, Wang, M, Sun, H. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of citrate, lactate and transferrin in determining titanium release from surgical devices into human serum.
J. Biol. Inorg. Chem., 23, 2018
|
|
7L1E
 
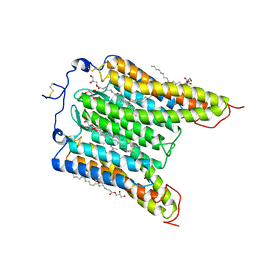 | | The Crystal Structure of Bromide-Bound GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin-1, BROMIDE ION, ... | | Authors: | Li, H, Huang, C.Y, Wang, M, Spudich, J.L, Zheng, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of bromide-bound Gt ACR1 reveals a pre-activated state in the transmembrane anion tunnel.
Elife, 10, 2021
|
|
7VBH
 
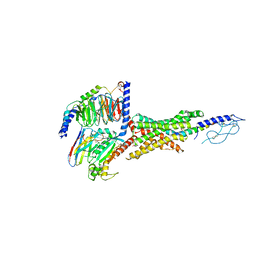 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7VAB
 
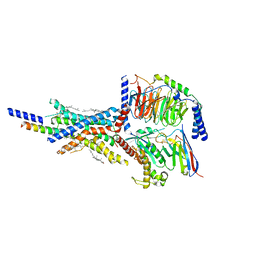 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-28 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
7E9V
 
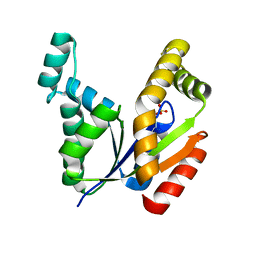 | | The Crystal Structure of human UMP-CMP kinase from Biortus. | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Wang, F, Lin, D, Wang, R, Wei, X, Shen, Z, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of human UMP-CMP kinase from Biortus.
To Be Published
|
|
6I5C
 
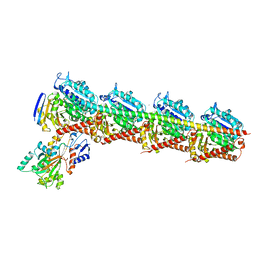 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
5N6L
 
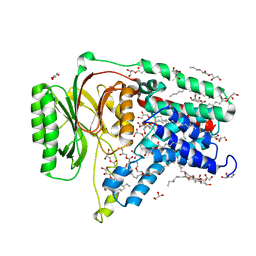 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
2D3U
 
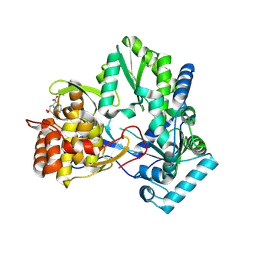 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
3U57
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
