7Q9W
 
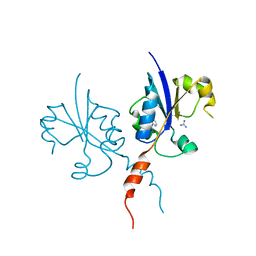 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 4-(aminomethyl)pyridin-2-amine | | Descriptor: | 4-(aminomethyl)pyridin-2-amine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QA7
 
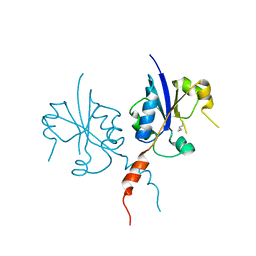 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Cyclopropanemethylamine | | Descriptor: | Cholinephosphate cytidylyltransferase, cyclopropylmethanamine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q9V
 
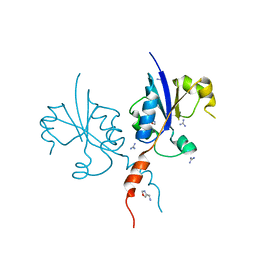 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-(1,3-oxazol-4-yl)methanamine | | Descriptor: | 1,3-oxazol-4-ylmethanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QAD
 
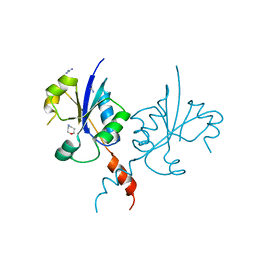 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1,4-Oxazepane hydrochloride | | Descriptor: | 1,4-oxazepane, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QD3
 
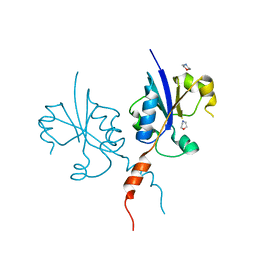 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with morpholine | | Descriptor: | Cholinephosphate cytidylyltransferase, morpholine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
5FTO
 
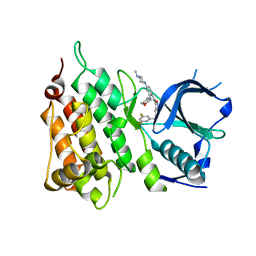 | | Crystal structure of the ALK kinase domain in complex with Entrectinib | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, Entrectinib | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
7QVO
 
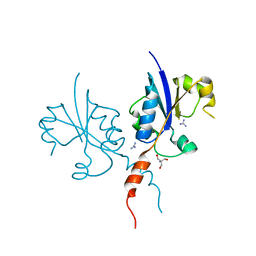 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with guanidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
5LWF
 
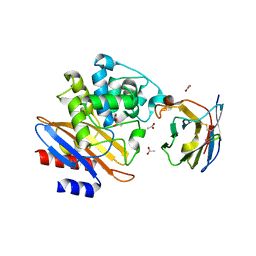 | | Structure of a single domain camelid antibody fragment cAb-G10S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | ACETATE ION, Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-G10S | | Authors: | Vettore, N, Kerff, F, Pain, C, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2016-09-16 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To Be Published
|
|
7QVN
 
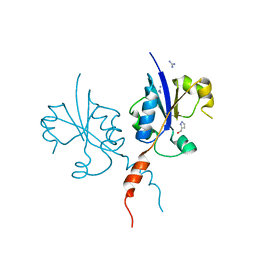 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with(1H-pyrazol-5-yl)methanol | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
3N9U
 
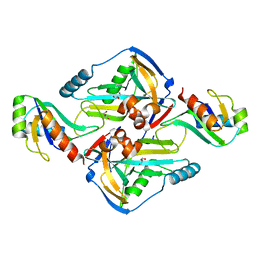 | | Crystal Structure of the Complex between the 25 kDa Subunit and the 59 kDa Subunit (RRM domain) of Human Cleavage Factor Im | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 7, GLYCEROL | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nielsen, T.N, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the Complex between the 25 kDa Subunit and the 59 kDa Subunit (RRM domain) of Human Cleavage Factor Im
To be Published
|
|
7QBJ
 
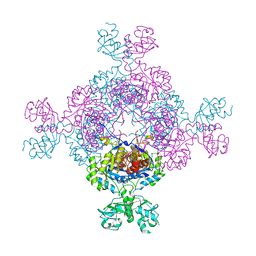 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QEM
 
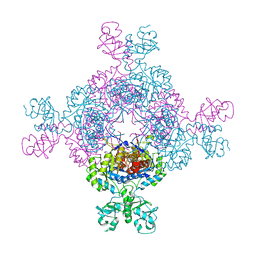 | | bacterial IMPDH chimera | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Gedeon, A, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2021-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QDX
 
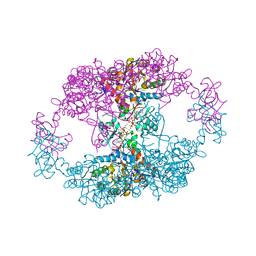 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
1RSV
 
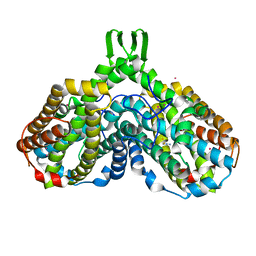 | | azide complex of the diferrous E238A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Assarsson, M, Andersson, M.E, Hogbom, M, Persson, B.O, Sahlin, M, Barra, A.L, Sjoberg, B.M, Nordlund, P, Graslund, A. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restoring proper radical generation by azide binding to the iron site of the E238A mutant R2 protein of ribonucleotide reductase from Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
3MTG
 
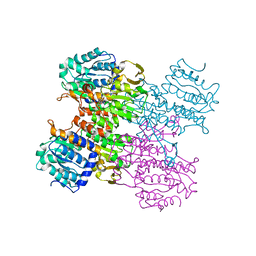 | | Crystal structure of human S-adenosyl homocysteine hydrolase-like 1 protein | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative adenosylhomocysteinase 2 | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of human S-adenosyl homocysteine hydrolase-like 1 protein
TO BE PUBLISHED
|
|
5AHM
 
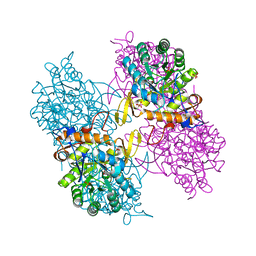 | | IMP-bound form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, PHOSPHATE ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
7R4K
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | MAGNESIUM ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4J
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | CALCIUM ION, NAD kinase 2, mitochondrial | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4L
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | FE (III) ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
7R4M
 
 | | Crystal structure of mitochondrial NAD kinase | | Descriptor: | CALCIUM ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
2Y6H
 
 | | X-2 L110F CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
5AQ9
 
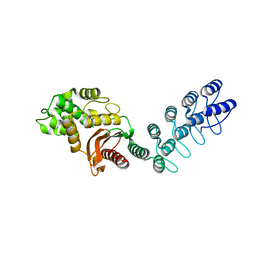 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, OFF7_DB08V4 | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQ8
 
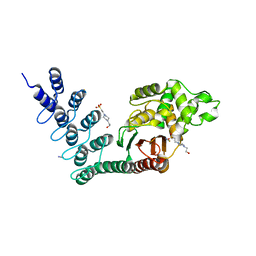 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, OFF7_DB12V4, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AQA
 
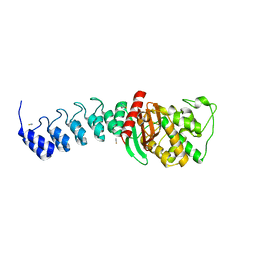 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | OFF7_DB04V3, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
