8WKS
 
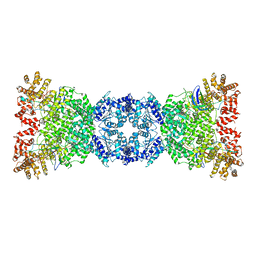 | | Cryo-EM structure of DSR2-TUBE complex | | Descriptor: | SIR2-like domain-containing protein, TUBE | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
3V6E
 
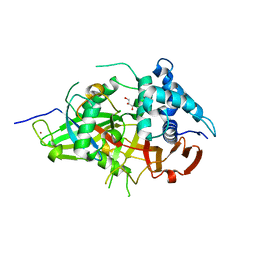 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
8DKN
 
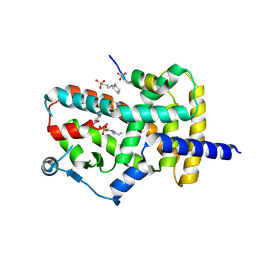 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
4GMB
 
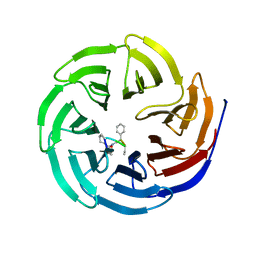 | | Crystal structure of human WD repeat domain 5 with compound MM-402 | | Descriptor: | MM-402, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J.Med.Chem., 60, 2017
|
|
3ADG
 
 | |
3ADJ
 
 | |
6BR1
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
7U8Z
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
6BRF
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BS2
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6Q32
 
 | |
6BRY
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
4I6L
 
 | |
3ADI
 
 | |
3ADL
 
 | |
4IVB
 
 | | JAK1 kinase (JH1 domain) in complex with the inhibitor TRANS-4-{2-[(1R)-1-HYDROXYETHYL]IMIDAZO[4,5-D]PYRROLO[2,3-B]PYRIDIN-1(6H)-YL}CYCLOHEXANECARBONITRILE | | Descriptor: | Tyrosine-protein kinase JAK1, trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexanecarbonitrile | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
8DKV
 
 | | PPARg bound to JTP-426467 and Co-R peptide | | Descriptor: | 2-chloro-N-[4-(5-methyl-1,3-benzoxazol-2-yl)phenyl]-5-nitrobenzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSY
 
 | | PPARg bound to inverse agonist H3B-343 | | Descriptor: | Peroxisome proliferator-activated receptor gamma, {5-[(5-{[(4-tert-butylphenyl)methyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}acetic acid | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
5HXL
 
 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
5D39
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ZDQ
 
 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (NUCLEOTIDE-FREE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, MITOCHONDRIAL, CARDIOLIPIN, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Krojer, T, von Delft, F, Vollmar, M, Mukhopadhyay, S, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6U4N
 
 | |
8GZL
 
 | |
