7MNM
 
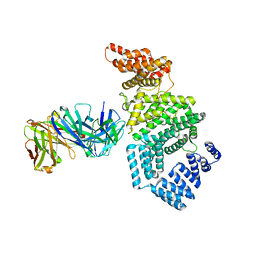 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T585M mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNQ
 
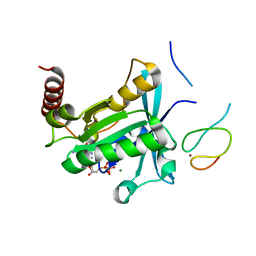 | | Crystal Structure of the ZnF2 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNT
 
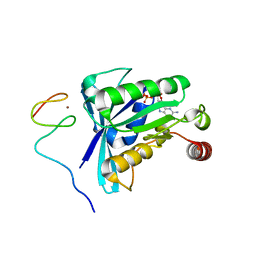 | | Crystal Structure of the ZnF5 or ZnF6 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNN
 
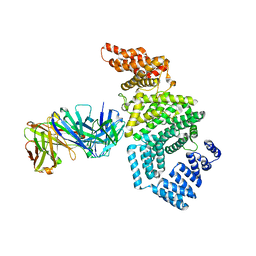 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T653I mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO3
 
 | | Crystal Structure of the ZnF3 of Nucleoporin NUP153 in complex with Ran-GDP, resolution 2.05 Angstrom | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MVU
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 1-1756; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW0
 
 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
3V69
 
 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5ZMY
 
 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
5ZCJ
 
 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
5ZMU
 
 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
2GYY
 
 | |
8HOM
 
 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
4RRS
 
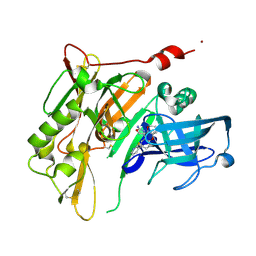 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'R,10a'S)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
8HOL
 
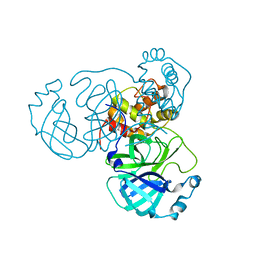 | |
8HOZ
 
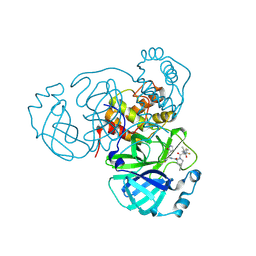 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
4RRO
 
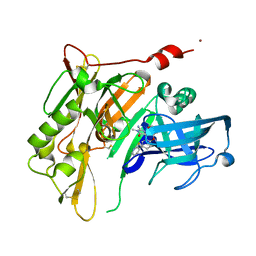 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RS4
 
 | |
2GZ1
 
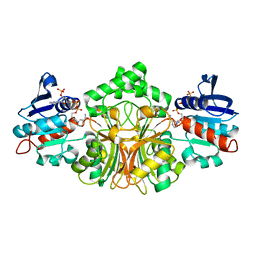 | | Structure of Aspartate Semialdehyde Dehydrogenase (ASADH) from Streptococcus pneumoniae complexed with NADP | | Descriptor: | Aspartate beta-semialdehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Faehnle, C.R, Le Coq, J, Liu, X, Viola, R.E. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examination of key intermediates in the catalytic cycle of aspartate-beta-semialdehyde dehydrogenase from a gram-positive infectious bacteria.
J.Biol.Chem., 281, 2006
|
|
4S1T
 
 | |
4RRN
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'S,10a'R)-2-amino-8'-(2-fluoropyridin-3-yl)-1-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4GO1
 
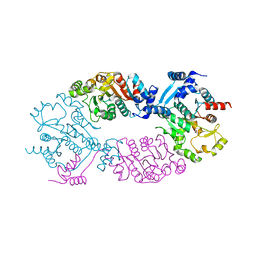 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
2L8S
 
 | |
