7TJD
 
 | |
7Y0L
 
 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7YD2
 
 | | SulE_P44R_S209A | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
5YWO
 
 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWF
 
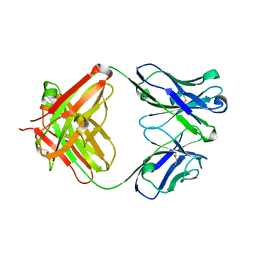 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
4FAD
 
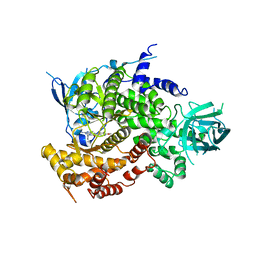 | | Design and Synthesis of a Novel Pyrrolidinyl Pyrido Pyrimidinone Derivative as a Potent Inhibitor of PI3Ka and mTOR | | Descriptor: | 2-amino-6-(5-fluoro-6-methoxypyridin-3-yl)-4-methyl-8-(pyrrolidin-1-yl)pyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Greasley, S.E, Knighton, D.R, LaFleur Rogers, C.M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of a novel pyrrolidinyl pyrido pyrimidinone derivative as a potent inhibitor of PI3Kalpha and mTOR
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5YWP
 
 | | JEV-2H4 Fab complex | | Descriptor: | 2H4 heavy chain, 2H4 light chain, JEV E protein, ... | | Authors: | Qiu, X.D, Lei, Y.F, Yang, P, Gao, Q, WANG, N, Cao, L, Yuan, S, Wang, X.X, Xu, Z.K, Rao, Z.H. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
7TOU
 
 | |
7TOV
 
 | |
7TOX
 
 | |
7TOY
 
 | |
7TP1
 
 | |
7TOZ
 
 | |
7TP0
 
 | |
7TP2
 
 | |
7TPH
 
 | |
7TP8
 
 | |
7TPA
 
 | |
7TPC
 
 | |
7TLA
 
 | |
7TPE
 
 | |
7TPF
 
 | |
7TPL
 
 | |
7TP9
 
 | |
7TLC
 
 | |
