6JGF
 
 | |
6JGV
 
 | |
6JGX
 
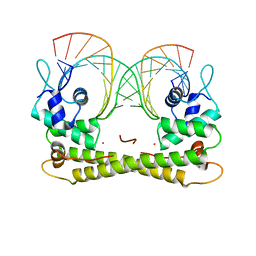 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JGW
 
 | |
6JNI
 
 | |
7EHE
 
 | | Acetolactate Synthase from Trichoderma harzianum | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zang, X, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
6J9O
 
 | |
6M6I
 
 | | Structure of HSV2 B-capsid portal vertex | | Descriptor: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
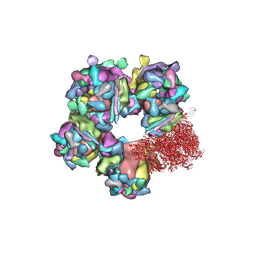 | | Structure of HSV2 C-capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
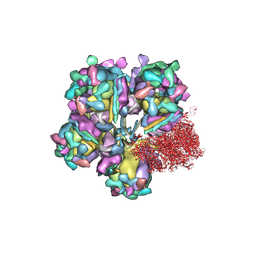 | | Structure of HSV2 viron capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.39 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
6A72
 
 | | Copper transporter protein | | Descriptor: | ATP7B protein, CALCIUM ION, dioxo(di-mu-sulfide)dimolybdenum | | Authors: | Chen, W.B. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrathiomolybdate induces dimerization of the metal-binding domain of ATPase and inhibits platination of the protein.
Nat Commun, 10, 2019
|
|
6A71
 
 | | Crystal Structure of Human ATP7B and TM Complex | | Descriptor: | ATP7B protein, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, W.B. | | Deposit date: | 2018-06-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tetrathiomolybdate induces dimerization of the metal-binding domain of ATPase and inhibits platination of the protein.
Nat Commun, 10, 2019
|
|
5UEH
 
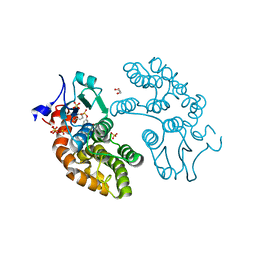 | | Structure of GSTO1 covalently conjugated to quinolinic acid fluorosulfate | | Descriptor: | 2-(4-chlorophenyl)-6-[(fluorosulfonyl)oxy]quinoline-4-carboxylic acid, GLYCEROL, Glutathione S-transferase omega-1, ... | | Authors: | Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
8JPX
 
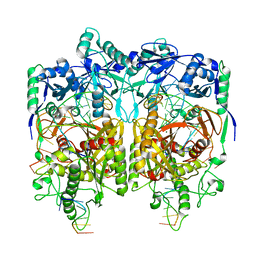 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
5T35
 
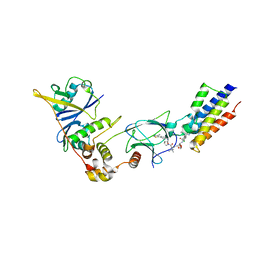 | | The PROTAC MZ1 in complex with the second bromodomain of Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Transcription elongation factor B polypeptide 1, ... | | Authors: | Gadd, M.S, Zengerle, M, Ciulli, A. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of PROTAC cooperative recognition for selective protein degradation.
Nat. Chem. Biol., 13, 2017
|
|
5UI4
 
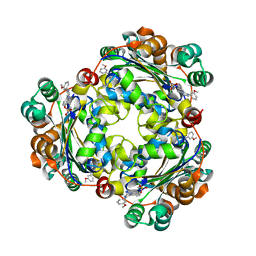 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | Descriptor: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | Authors: | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
3KVX
 
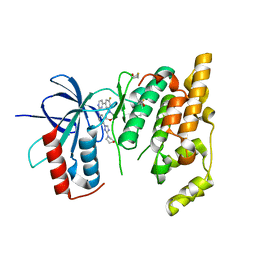 | | JNK3 bound to aminopyrimidine inhibitor, SR-3562 | | Descriptor: | Mitogen-activated protein kinase 10, N-[(2Z)-4-(3-fluoro-5-morpholin-4-ylphenyl)pyrimidin-2(1H)-ylidene]-4-(3-morpholin-4-yl-1H-1,2,4-triazol-1-yl)aniline | | Authors: | Habel, J.E, Laughlin, J.D, LoGrasso, P. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors
J.Med.Chem., 53, 2010
|
|
6IMR
 
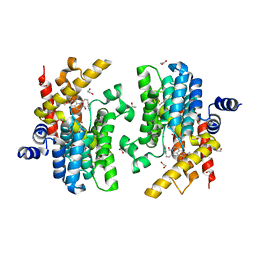 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[3-(1H-indol-3-yl)propyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMB
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6INK
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMD
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IND
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-6,7-dimethoxy-1-[2-(6-methyl-1H-indol-3-yl)ethyl]-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3FI3
 
 | | Crystal structure of JNK3 with indazole inhibitor, SR-3737 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(2-fluorophenyl)amino]-1H-indazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E, Duckett, D, LoGrasso, P. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
