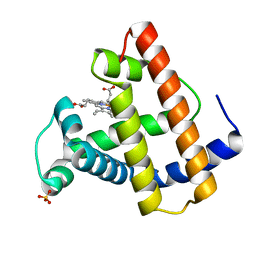
1MOA
 
 | |
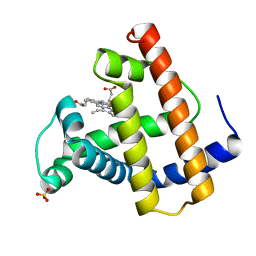
1MOB
 
 | |
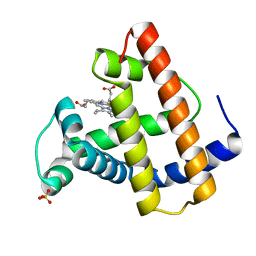
1MOD
 
 | |
4QIR
 
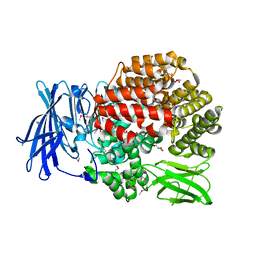 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | Descriptor: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | Deposit date: | 2014-06-01 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QHP
 
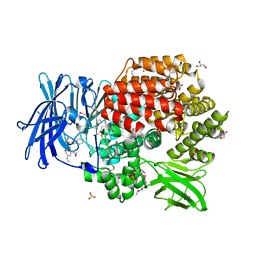 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe(4-CH2NH2) | | Descriptor: | (2R)-2-[4-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, (2S)-2-[4-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, Aminopeptidase N, ... | | Authors: | Nocek, B, Joachimiak, A. | | Deposit date: | 2014-05-28 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
2R0L
 
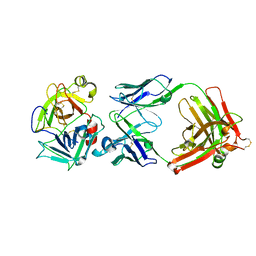 | | Short Form HGFA with Inhibitory Fab75 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TJO
 
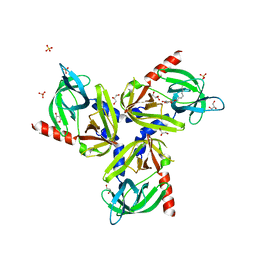 | |
3TJN
 
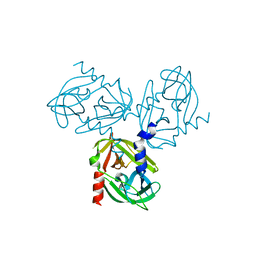 | | HtrA1 catalytic domain, apo form | | Descriptor: | Serine protease HTRA1 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of HtrA1 and Its Subdomains.
Structure, 20, 2012
|
|
2R0K
 
 | | Protease domain of HGFA with inhibitor Fab58 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain of Fab58, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TJQ
 
 | | N-domain of HtrA1 | | Descriptor: | CHLORIDE ION, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and Functional Analysis of HtrA1 and Its Subdomains.
Structure, 20, 2012
|
|
3EER
 
 | | High resolution structure of putative organic hydroperoxide resistance protein from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, IMIDAZOLE, Organic hydroperoxide resistance protein, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-05 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the organic hydroperoxide
resistance protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
1HVX
 
 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
5TCZ
 
 | |
3LFT
 
 | |
1FHV
 
 | |
3R5X
 
 | | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-20 | | Release date: | 2011-04-06 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of D-alanine--D-Alanine Ligase from Bacillus anthracis complexed with ATP
To be Published
|
|
1FHU
 
 | |
1NMV
 
 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NMW
 
 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1Q5X
 
 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
