8A67
 
 | |
8SY4
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-SER-LEU-ALA-SNC-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-05-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8SW2
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETATE ION, LEU-AIB-ALA-SER-LEU-ALA-CYS-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-05-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
7ZJ4
 
 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
8A58
 
 | |
8A9P
 
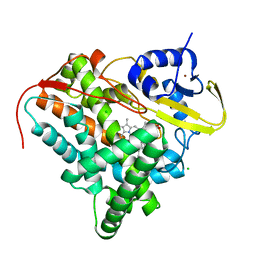 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
7ZJ5
 
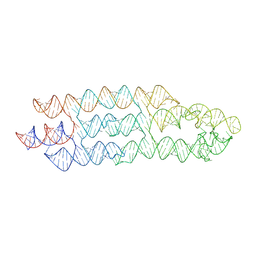 | | Unbound state of a brocolli-pepper aptamer FRET tile. | | Descriptor: | POTASSIUM ION, brocolli-pepper aptamer | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7ZM5
 
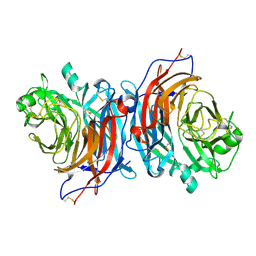 | | Structure of Mossman virus receptor binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Stelfox, A.J, Bowden, T.A, Rissanen, I, Harlos, K. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
7ZJV
 
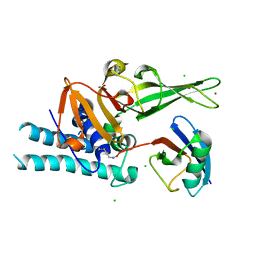 | |
8SP7
 
 | |
8TSZ
 
 | |
8TSX
 
 | |
8TSU
 
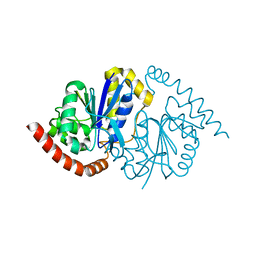 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT0
 
 | |
8TT1
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSY
 
 | |
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TWI
 
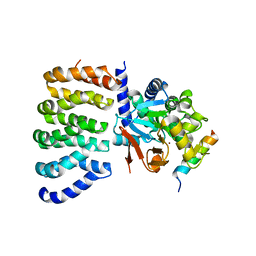 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8TWE
 
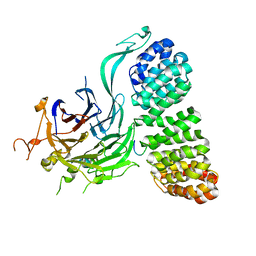 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body | | Descriptor: | PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8PW7
 
 | | A respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
8PW5
 
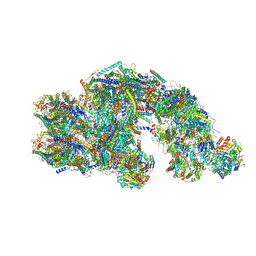 | | CS respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
8PW6
 
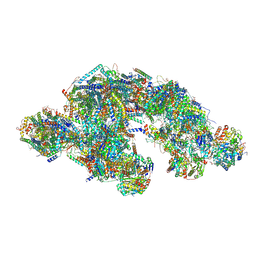 | | C respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 2024
|
|
8PVZ
 
 | |
