1WDF
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
118D
 
 | |
1WDG
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4YLY
 
 | | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, staphylococcus aureus at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
1XXE
 
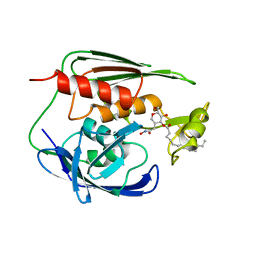 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
1Y4E
 
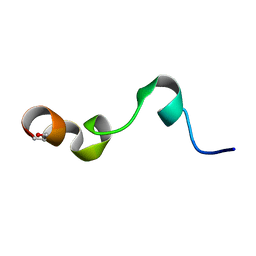 | | NMR structure of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Slepkov, E.R, Rainey, J.K, Li, X, Liu, Y, Lindhout, D.A, Sykes, B.D, Fliegel, L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger.
J.Biol.Chem., 280, 2005
|
|
8IGT
 
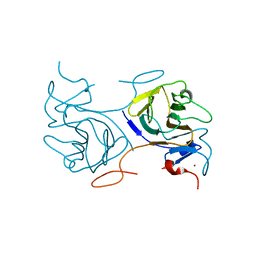 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A1 | | Descriptor: | Butyrophilin subfamily 2 member A1, ZINC ION | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-21 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
4LOG
 
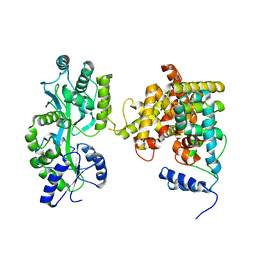 | | The crystal structure of the orphan nuclear receptor PNR ligand binding domain fused with MBP | | Descriptor: | Maltose ABC transporter periplasmic protein and NR2E3 protein chimeric construct | | Authors: | Tan, M.E, Zhou, X.E, Soon, F.-F, Li, X, Li, J, Yong, E.-L, Melcher, K, Xu, H.E. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Orphan Nuclear Receptor NR2E3/PNR Ligand Binding Domain Reveals a Dimeric Auto-Repressed Conformation.
Plos One, 8, 2013
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6XBW
 
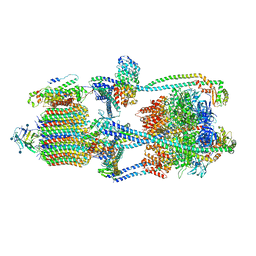 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
1QST
 
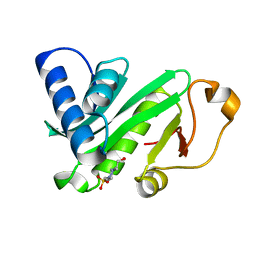 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
6XBY
 
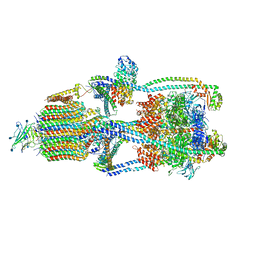 | | Cryo-EM structure of V-ATPase from bovine brain, state 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
8JYE
 
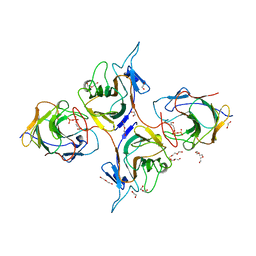 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYC
 
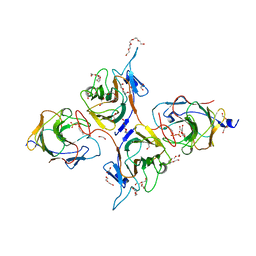 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
1QSR
 
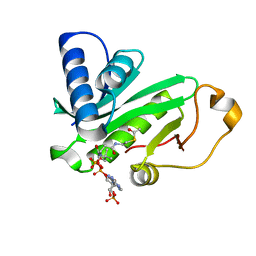 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
6XE6
 
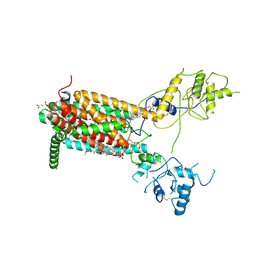 | | Structure of Human Dispatched-1 (DISP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Chen, H, Liu, Y, Li, X. | | Deposit date: | 2020-06-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure of human Dispatched-1 provides insights into Hedgehog ligand biogenesis.
Life Sci Alliance, 3, 2020
|
|
7Y9O
 
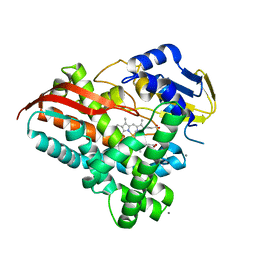 | | Crystal structure of a CYP109B4 variant from Bacillus sonorensis | | Descriptor: | CALCIUM ION, Cytochrome P450 monooxygenase YjiB, IMIDAZOLE, ... | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
7Y97
 
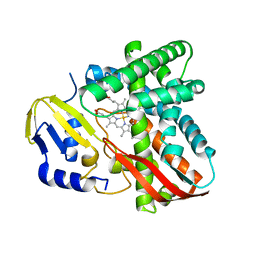 | | Crystal structure of CYP109B4 from Bacillus Sonorensis | | Descriptor: | Cytochrome P450 monooxygenase YjiB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
7Y98
 
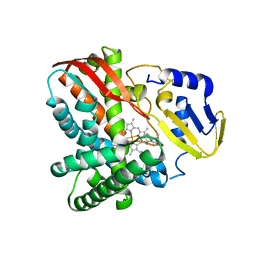 | | Crystal structure of CYP109B4 from Bacillus Sonorensis in complex with Testosterone | | Descriptor: | Cytochrome P450 monooxygenase YjiB, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
8U3E
 
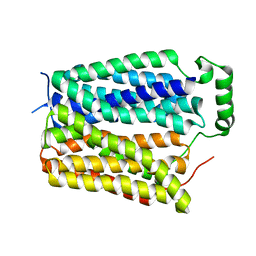 | | Structure of Apo Sialin at pH5.0 | | Descriptor: | Sialin | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and inhibition of the human lysosomal transporter Sialin.
Nat Commun, 15, 2024
|
|
8U3D
 
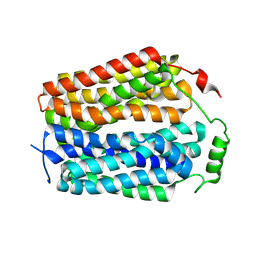 | | Structure of Apo Sialin at pH7.5 | | Descriptor: | Sialin | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure and inhibition of the human lysosomal transporter Sialin.
Nat Commun, 15, 2024
|
|
8U3F
 
 | | Structure of Apo Sialin R168K mutant | | Descriptor: | Sialin | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and inhibition of the human lysosomal transporter Sialin.
Nat Commun, 15, 2024
|
|
