2MVM
 
 | | Solution structure of eEF1Bdelta CAR domain | | Descriptor: | Elongation factor 1-delta | | Authors: | Wu, H, Feng, Y. | | Deposit date: | 2014-10-09 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolutionarily Conserved Binding of Translationally Controlled Tumor Protein to Eukaryotic Elongation Factor 1B.
J.Biol.Chem., 290, 2015
|
|
7C78
 
 | |
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
2MVN
 
 | |
2KQ3
 
 | | Solution structure of SNase140 | | Descriptor: | Thermonuclease | | Authors: | Wang, M, Feng, Y, Yao, H, Wang, J. | | Deposit date: | 2009-10-26 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Importance of the C-Terminal Loop L137-S141 for the Folding and Folding Stability of Staphylococcal Nuclease
Biochemistry, 49, 2010
|
|
2LCC
 
 | | Solution structure of RBBP1 chromobarrel domain | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Gong, W, Feng, Y. | | Deposit date: | 2011-04-28 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of methylated histone tails by retinoblastoma-binding protein 1.
J.Biol.Chem., 2012
|
|
7XYB
 
 | | The cryo-EM structure of an AlpA-loaded complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XM0
 
 | |
7CG5
 
 | |
7CG8
 
 | | Structure of the sensor domain (short construct) of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, Anti-sigma factor RsgI, ... | | Authors: | Dong, S, Feng, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sensor domain of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens
To Be Published
|
|
7CG1
 
 | |
7DM4
 
 | | Solution structure of ARID4B Tudor domain | | Descriptor: | AT-rich interactive domain-containing protein 4B | | Authors: | Ren, J, Yao, H, Hu, W, Perrett, S, Feng, Y, Gong, W. | | Deposit date: | 2020-12-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the DNA-binding activity of human ARID4B Tudor domain.
J.Biol.Chem., 296, 2021
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7ELE
 
 | |
7ELD
 
 | |
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
4KFW
 
 | |
4KFV
 
 | |
6OAH
 
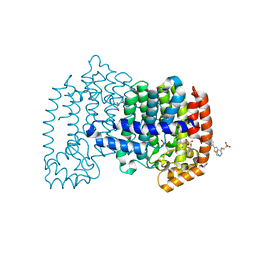 | |
6OAG
 
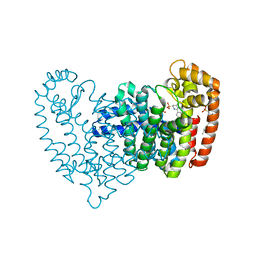 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
7Y67
 
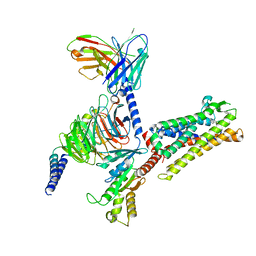 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | Descriptor: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y65
 
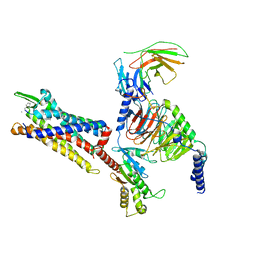 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | Descriptor: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
6N82
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02037 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Farnesyl pyrophosphate synthase, ... | | Authors: | Park, J, Schilling, M.A, Berghuis, A.M. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
