7Y0F
 
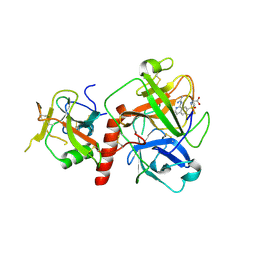 | | Crystal structure of TMPRSS2 in complex with UK-371804 | | Descriptor: | 2-[(1-carbamimidamido-4-chloranyl-isoquinolin-7-yl)sulfonylamino]-2-methyl-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0E
 
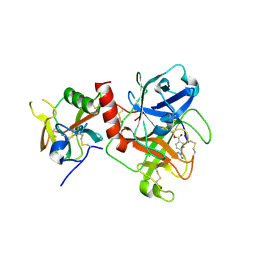 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8YXW
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCC-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
5XKA
 
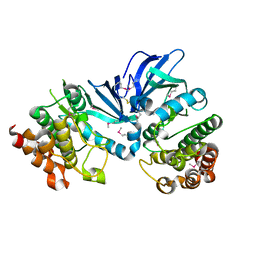 | | Crystal structure of M.tuberculosis PknI kinase domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Yan, Q, Jiang, D, Qian, L, Zhang, Q, Zhang, W, Zhou, W, Mi, K, Guddat, L, Yang, H, Rao, Z. | | Deposit date: | 2017-05-06 | | Release date: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5ZHZ
 
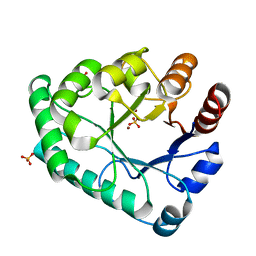 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
8PUH
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PU6
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PU7
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUA
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PU9
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUB
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUC
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUD
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PU8
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUF
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUE
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
7WIX
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIV
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIU
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIW
 
 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB complexed with ATP in an occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
