6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
5F1C
 
 | | Crystal structure of an invertebrate P2X receptor from the Gulf Coast tick in the presence of ATP and Zn2+ ion at 2.9 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, ... | | Authors: | Kasuya, G, Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels
Cell Rep, 14, 2016
|
|
3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
3WQH
 
 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | Authors: | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | Deposit date: | 2014-01-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
6IU4
 
 | | Crystal structure of iron transporter VIT1 with cobalt ion | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU3
 
 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
3CAF
 
 | |
7DPA
 
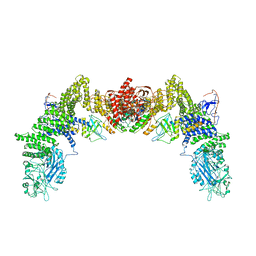 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
3CU1
 
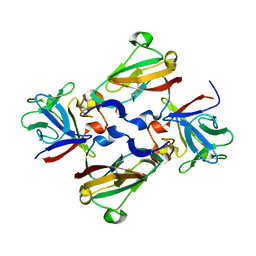 | | Crystal Structure of 2:2:2 FGFR2D2:FGF1:SOS complex | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor receptor 2, Heparin-binding growth factor 1 | | Authors: | Guo, F, Dakshinamurthy, R, Thallapuranam, S.K.K, Sakon, J. | | Deposit date: | 2008-04-15 | | Release date: | 2009-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of 2:2:2 FGFR2D2:FGF1:SOS complex
To be Published
|
|
3VQV
 
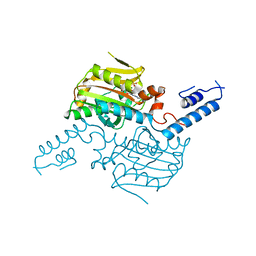 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
6IJE
 
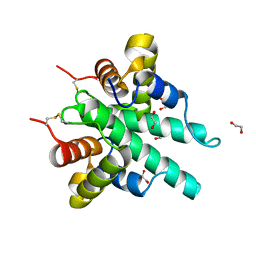 | | Crystal structure of the type VI amidase immunity (Tai4) from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, Tai4 | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6IJF
 
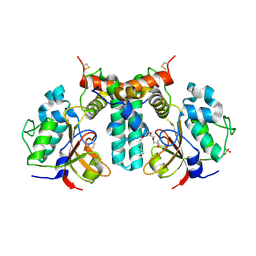 | | Crystal structure of the type VI effector-immunity complex (Tae4-Tai4) from Agrobacterium tumefaciens | | Descriptor: | PENTAETHYLENE GLYCOL, SULFATE ION, Tae4, ... | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3WXM
 
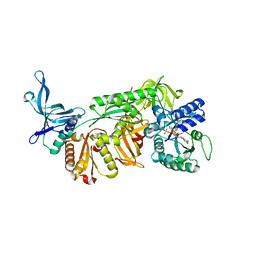 | | Crystal structure of archaeal Pelota and GTP-bound EF1 alpha complex | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1 alpha complex
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6LX4
 
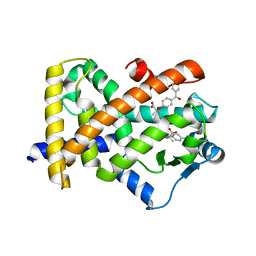 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
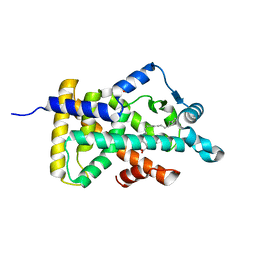 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
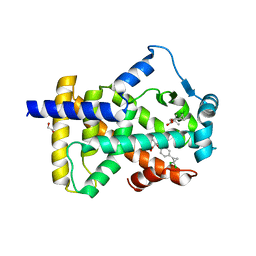 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX6
 
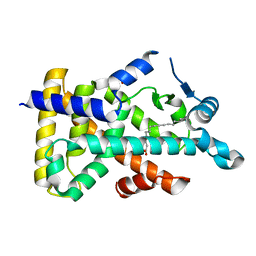 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXB
 
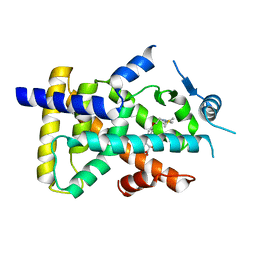 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6IU9
 
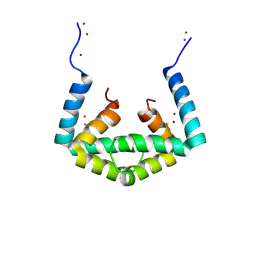 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
5GVS
 
 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
6LXA
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX8
 
 | | X-ray structure of human PPARalpha ligand binding domain-oleic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, OLEIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5GVR
 
 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
6IU8
 
 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
