7W3D
 
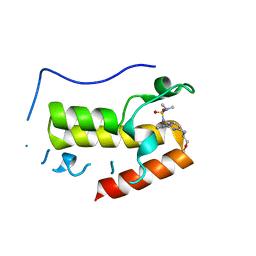 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
6JN8
 
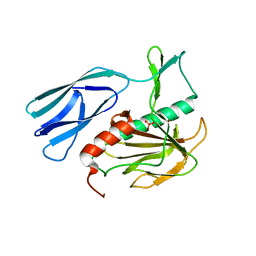 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
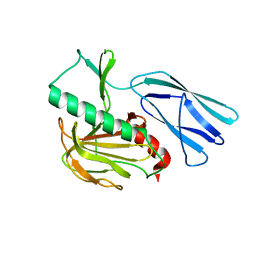 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
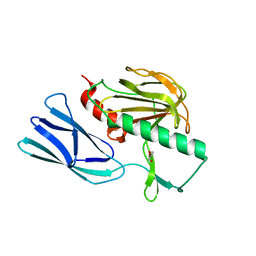 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
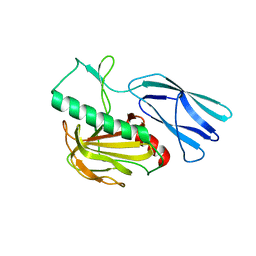 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
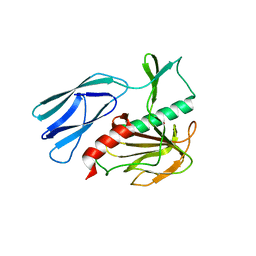 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
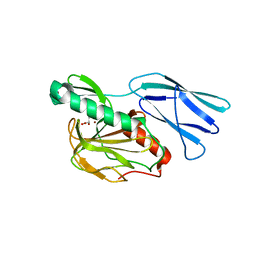 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
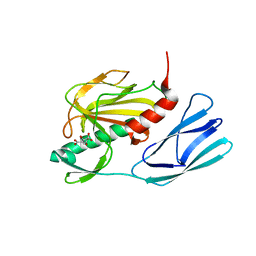 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
2J7Z
 
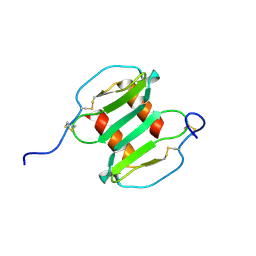 | | Crystal Structure of recombinant Human Stromal Cell-Derived Factor- 1alpha | | Descriptor: | STROMAL CELL-DERIVED FACTOR 1 ALPHA | | Authors: | Ryu, E.K, Kim, T.G, Kwon, T.H, Jung, I.D, Ryu, D.W, Park, Y.-M, Ahn, K, Ban, C. | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Recombinant Human Stromal Cell-Derived Factor-1Alpha.
Proteins, 67, 2007
|
|
3SXF
 
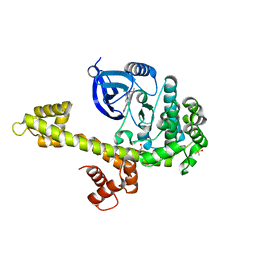 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-89 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-14 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T3V
 
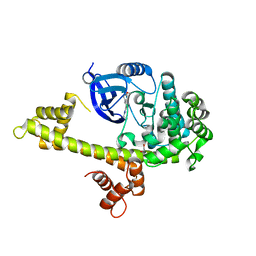 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-87 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T3U
 
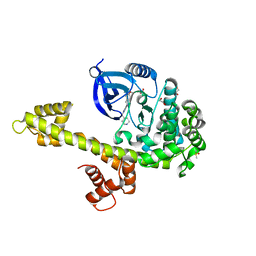 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-130 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
4O9C
 
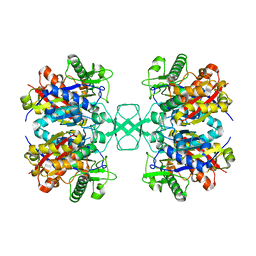 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | Descriptor: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poulos, T.L, Kim, J, Murarka, V. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.40000689 Å) | | Cite: | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
6MFJ
 
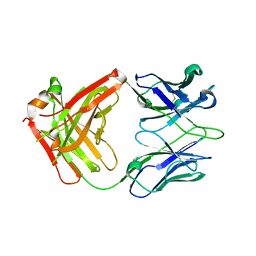 | |
3ZDQ
 
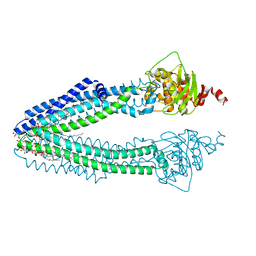 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (NUCLEOTIDE-FREE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, MITOCHONDRIAL, CARDIOLIPIN, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Krojer, T, von Delft, F, Vollmar, M, Mukhopadhyay, S, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MFP
 
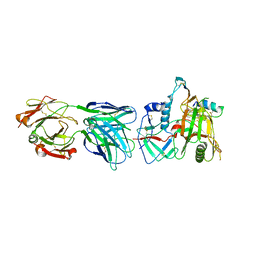 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
2OO8
 
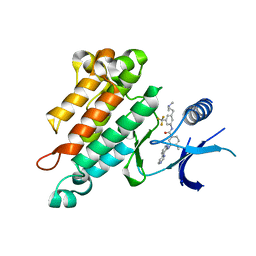 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5UIX
 
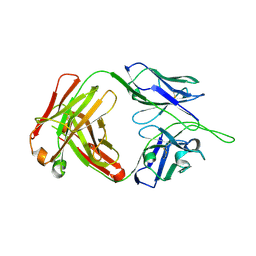 | |
2OSC
 
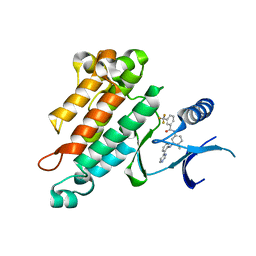 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6BFA
 
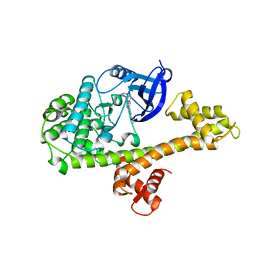 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1553 | | Descriptor: | 1-{4-amino-3-[6-(cyclopropyloxy)naphthalen-2-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
3SX9
 
 | |
1KOZ
 
 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | Descriptor: | Voltage-dependent Channel Inhibitor | | Authors: | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | Deposit date: | 2001-12-25 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
4NFX
 
 | |
4NFW
 
 | | Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli | | Descriptor: | MANGANESE (II) ION, Putative Nudix hydrolase ymfB, SULFATE ION | | Authors: | Hong, M.K, Kim, J.K, Kang, L.W. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-14 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divalent metal ion-based catalytic mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
