7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
4EHQ
 
 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
6ZGP
 
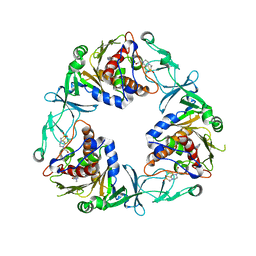 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with inhibitor MMV12 (MMV020670) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D.H, Cameron, A, Chen, Y. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
7V7M
 
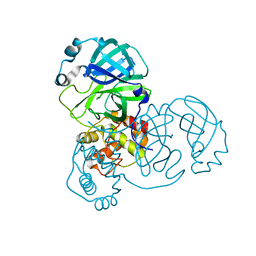 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
7VMB
 
 | |
1ZGK
 
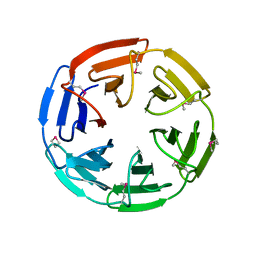 | | 1.35 angstrom structure of the Kelch domain of Keap1 | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Li, X, Bottoms, C.A, Hannink, M, Beamer, L.J. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1F95
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
5J6P
 
 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
6LAD
 
 | |
6LAF
 
 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | Descriptor: | Amuc_1100, SULFATE ION | | Authors: | Wang, J, Xiang, R, Zhang, M, Wang, M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
1M5Z
 
 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
7V52
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V57
 
 | | Structure of AdaV | | Descriptor: | 2-OXOGLUTARIC ACID, AdaV, CHLORIDE ION, ... | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V54
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V56
 
 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7V7X
 
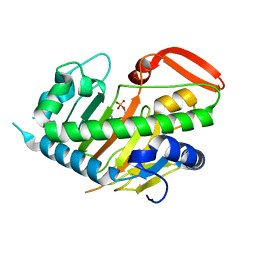 | | Structure of H194A AdaV | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, AdaV | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
1F96
 
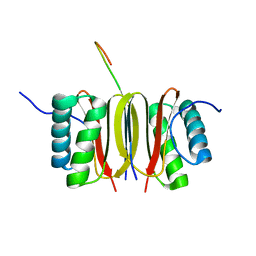 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | Descriptor: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
6GL8
 
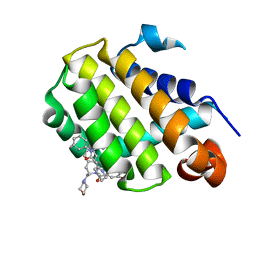 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
1B8Q
 
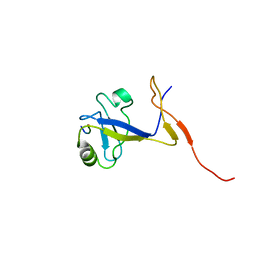 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
