3FG8
 
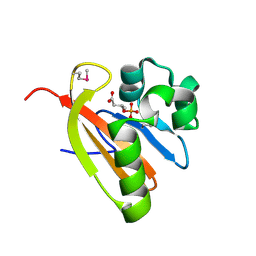 | | Crystal structure of PAS domain of RHA05790 | | Descriptor: | (3R)-3-(phosphonooxy)butanoic acid, uncharacterized protein RHA05790 | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PAS domain of RHA05790
To be Published
|
|
3D6J
 
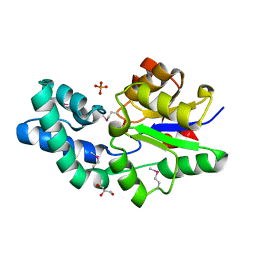 | | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative haloacid dehalogenase-like hydrolase | | Authors: | Chang, C, Wu, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis
To be Published
|
|
3COL
 
 | |
3FF4
 
 | |
4LMI
 
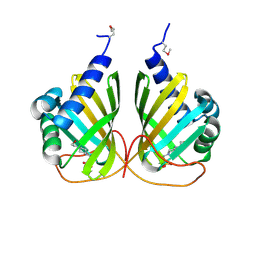 | |
3F42
 
 | | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, protein HP0035 | | Authors: | Chang, C, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori
To be Published
|
|
4MLZ
 
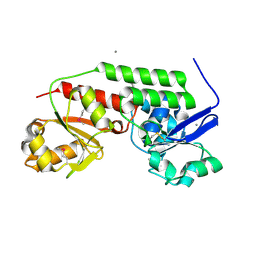 | | Crystal structure of periplasmic binding protein from Jonesia denitrificans | | Descriptor: | CALCIUM ION, POTASSIUM ION, Periplasmic binding protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of periplasmic binding protein from Jonesia denitrificans
To be Published
|
|
3D1L
 
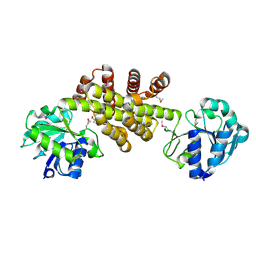 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | Descriptor: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | Authors: | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
4MDY
 
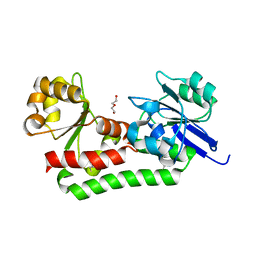 | |
4MJD
 
 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | Ketosteroid isomerase fold protein Hmuk_0747, MAGNESIUM ION, SODIUM ION | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3EYT
 
 | | Crystal structure of Thioredoxin-like superfamily protein SPOA0173 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, uncharacterized protein SPOA0173 | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Thioredoxin-like superfamily protein SPOA0173
To be Published
|
|
3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
4MOZ
 
 | |
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
4LQB
 
 | | Crystal structure of uncharacterized protein Kfla3161 | | Descriptor: | CITRIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of uncharacterized protein Kfla3161
To be Published
|
|
4MQD
 
 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
3E0K
 
 | |
3EFA
 
 | | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chang, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum
To be Published
|
|
3FWZ
 
 | | Crystal structure of TrkA-N domain of inner membrane protein ybaL from Escherichia coli | | Descriptor: | ADENOSINE MONOPHOSPHATE, Inner membrane protein ybaL, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of TrkA-N domain of inner membrane protein ybaL from Escherichia coli
To be Published
|
|
4O2I
 
 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
3GDW
 
 | | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, Sigma-54 interaction domain protein | | Authors: | Chang, C, Sather, A, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis
To be Published
|
|
3DED
 
 | | C-terminal domain of Probable hemolysin from Chromobacterium violaceum | | Descriptor: | CALCIUM ION, Probable hemolysin | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of C-terminal domain of Probable hemolysin from Chromobacterium violaceum
To be Published
|
|
3GDZ
 
 | | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Arginyl-tRNA synthetase | | Authors: | Chang, C, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of arginyl-tRNA synthetase from Klebsiella pneumoniae subsp. pneumoniae
To be Published
|
|
