7TMU
 
 | | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis
To Be Published
|
|
7TRW
 
 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | Authors: | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
7TKV
 
 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
7TJ1
 
 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-14 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
7TVX
 
 | |
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6X4I
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | Descriptor: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | Authors: | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WTC
 
 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6X9Y
 
 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WXC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
4IQE
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
4M8L
 
 | | crystal structure of RpiA-R5P complex | | Descriptor: | CHLORIDE ION, FORMAMIDE, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Rostankowski, R, Borek, D, Orlikowska, M, Nakka, C, Grimshaw, S, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | crystal structure of RpiA-R5P complex
TO BE PUBLISHED
|
|
4QM1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
6N36
 
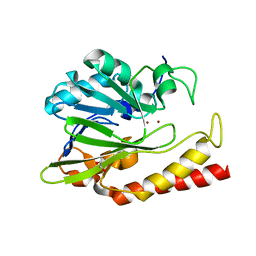 | | Beta-lactamase from Chitinophaga pinensis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Beta-lactamase from Chitinophaga pinensis
to be published
|
|
6N7F
 
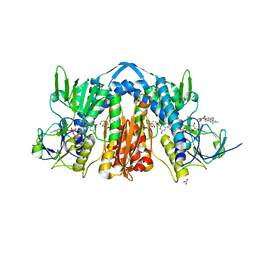 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
6N9K
 
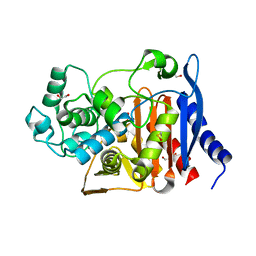 | | Beta-lactamase from Escherichia coli str. Sakai | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Beta-lactamase from Escherichia coli str. Sakai
to be published
|
|
6N1N
 
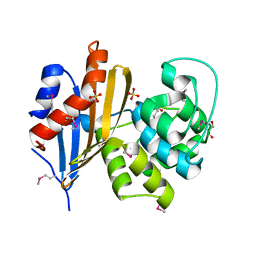 | | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Michalska, K, Tesar, C, Endres, M, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-09 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386
To Be Published
|
|
6N0I
 
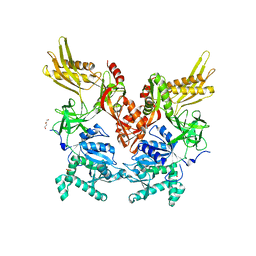 | | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor G 2, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.
To Be Published
|
|
6MU9
 
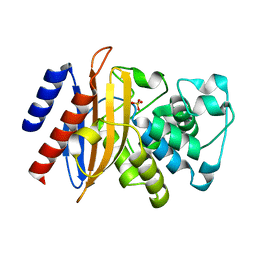 | |
6N7M
 
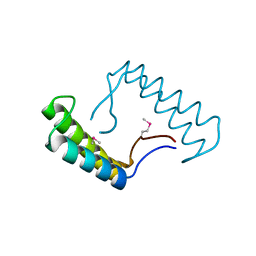 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | Descriptor: | Hypothetical Protein CD630_05490 | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
6NFP
 
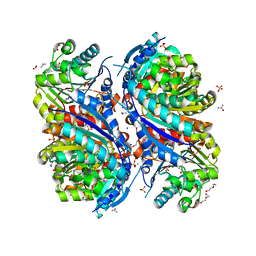 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
