5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
2WPT
 
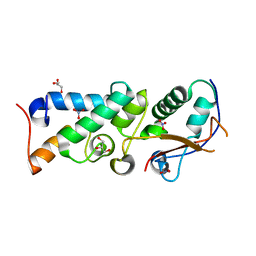 | | The crystal structure of Im2 in complex with colicin E9 DNase | | Descriptor: | COLICIN-E2 IMMUNITY PROTEIN, COLICIN-E9, GLYCEROL, ... | | Authors: | Meenan, N.A, Sharma, A, Fleishman, S.J, Macdonald, C.J, Boetzel, R, Moore, G.R, Baker, D, Kleanthous, C. | | Deposit date: | 2009-08-10 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Structural and Energetic Basis for High Selectivity in a High-Affinity Protein-Protein Interaction.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3GEQ
 
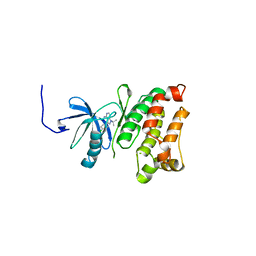 | | Structural basis for the chemical rescue of Src kinase activity | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
5W9F
 
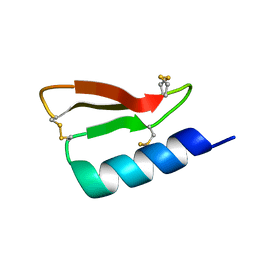 | | Solution structure of the de novo mini protein gHEEE_02 | | Descriptor: | De novo mini protein gHEEE_02 | | Authors: | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | Deposit date: | 2017-06-23 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
5UOI
 
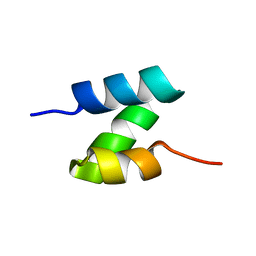 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | Descriptor: | HHH_rd1_0142 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
7M5T
 
 | |
7M0Q
 
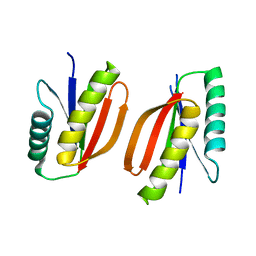 | |
7MWQ
 
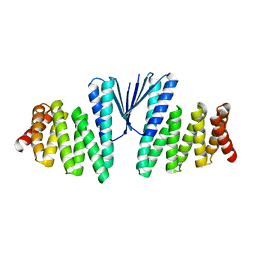 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | Descriptor: | LHD29A53, LHD29B53 | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWR
 
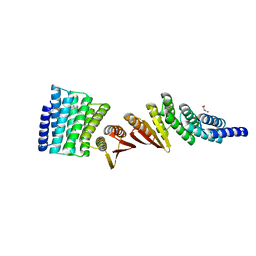 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | Descriptor: | LHD101A54, LHD101B4, MALONATE ION | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
4HHU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR280. | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, OR280, TETRAETHYLENE GLYCOL | | Authors: | Vorobiev, S, Lew, S, Lin, Y.-R, Seetharaman, J, Castelllanos, J, Maglaqui, M, Xiao, R, Lee, D, Koga, N, Koga, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-10 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein OR280.
To be Published
|
|
4HQD
 
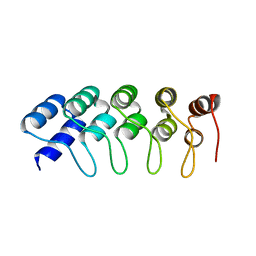 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR265. | | Descriptor: | Engineered Protein OR265 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal Structure of Engineered Protein OR265.
To be Published
|
|
4HXT
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR329 | | Descriptor: | De Novo Protein OR329 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, X, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
7RMX
 
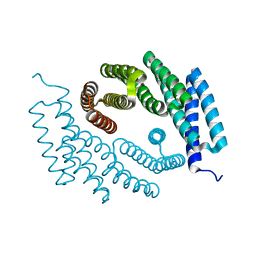 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4JXI
 
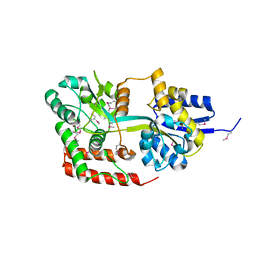 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
4JBC
 
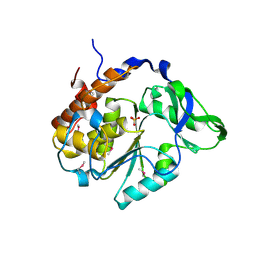 | | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318 | | Descriptor: | PHOSPHATE ION, designed serine hydrolase 3mmj_2 | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Baker, D, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318
To be Published
|
|
4JGK
 
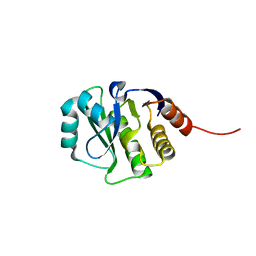 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
4K0C
 
 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
4IJB
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR288 | | Descriptor: | Engineered Protein OR288 | | Authors: | Vorobiev, S, Su, M, Bick, M.J, Seetharaman, J, Khare, S, Maglaqui, M, Xiao, R, Lee, D, Day, A, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Crystal Structure of Engineered Protein OR288.
To be Published
|
|
4J4Z
 
 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
4KYB
 
 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
2VLP
 
 | | R54A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
