1M4B
 
 | | Crystal Structure of Human Interleukin-2 K43C Covalently Modified at C43 with 2-[2-(2-Cyclohexyl-2-guanidino-acetylamino)-acetylamino]-N-(3-mercapto-propyl)-propionamide | | Descriptor: | 2-[2-(2-CYCLOHEXYL-2-GUANIDINO-ACETYLAMINO)-ACETYLAMINO]-N-(3-MERCAPTO-PROPYL)-PROPIONAMIDE, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M48
 
 | | Crystal Structure of Human IL-2 Complexed with (R)-N-[2-[1-(Aminoiminomethyl)-3-piperidinyl]-1-oxoethyl]-4-(phenylethynyl)-L-phenylalanine methyl ester | | Descriptor: | 2-[3-METHYL-4-(N-METHYL-GUANIDINO)-BUTYRYLAMINO]-3-(4-PHENYLETHYNYL-PHENYL)-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M47
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | SULFATE ION, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, Wells, J.A, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Binding of small molecules to an adaptive protein-protein interface.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M4C
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M49
 
 | | Crystal Structure of Human Interleukin-2 Complexed with SP-1985 | | Descriptor: | 2-[2-(1-CARBAMIMIDOYL-PIPERIDIN-3-YL)-ACETYLAMINO]-3-{4-[2-(3-OXALYL-1H-INDOL-7-YL)ETHYL]-PHENYL}-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M4A
 
 | | Crystal Structure of Human Interleukin-2 Y31C Covalently Modified at C31 with (1H-Indol-3-yl)-(2-mercapto-ethoxyimino)-acetic acid | | Descriptor: | (1H-INDOL-3-YL)-(2-MERCAPTO-ETHOXYIMINO)-ACETIC ACID, GLYCEROL, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2QD0
 
 | | Crystal structure of mitoNEET | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Lin, J, Zhou, T, Ye, K, Wang, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8D4P
 
 | |
3KXT
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA complex | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', Chromatin protein Cren7 | | Authors: | Feng, Y, Wang, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex
Protein Sci., 19, 2010
|
|
4DTX
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | CALCIUM ION, DNA polymerase, DNA primer, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4E3S
 
 | | RB69 DNA Polymerase Ternary Complex with dQTP Opposite dT | | Descriptor: | 3-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-7-methyl-3H-imidazo[4,5-b]pyridine, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-03-10 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DWR
 
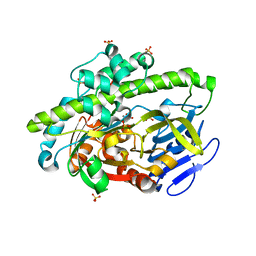 | | RNA ligase RtcB/Mn2+ complex | | Descriptor: | MANGANESE (II) ION, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Xia, S, Englert, M, Soll, D, Wang, J. | | Deposit date: | 2012-02-26 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1MIV
 
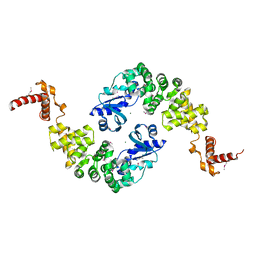 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
9IUZ
 
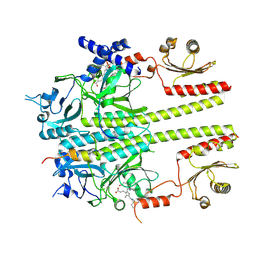 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MHK
 
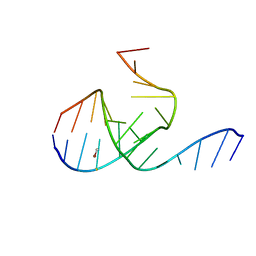 | | Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn | | Descriptor: | BROMIDE ION, RNA 12-mer BCh12, RNA 14-mer BCh12 | | Authors: | Szep, S, Wang, J, Moore, P.B. | | Deposit date: | 2002-08-20 | | Release date: | 2002-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a 26-nucleotide RNA containing a hook-turn
RNA, 9, 2003
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
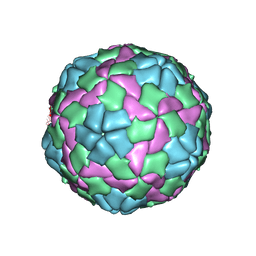 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5EFI
 
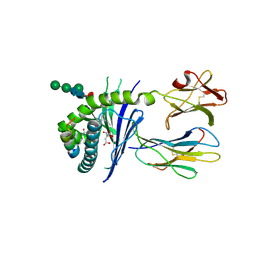 | | Crystal structure of mouse CD1d in complex with the p99p lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an alpha-Helical Peptide and Lipopeptide Bound to the Nonclassical Major Histocompatibility Complex (MHC) Class I Molecule CD1d.
J.Biol.Chem., 291, 2016
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
8ZH5
 
 | |
6KRX
 
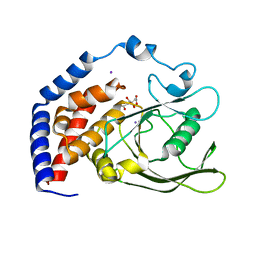 | | Crystal Structure of AtPTP1 at 1.7 angstrom | | Descriptor: | CITRATE ANION, IODIDE ION, Protein-tyrosine-phosphatase PTP1 | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AtPTP1 positively mediates brassinosteroid signaling from receptor kinases to GSK3-like kinase BIN2
To Be Published
|
|
6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
