8GSS
 
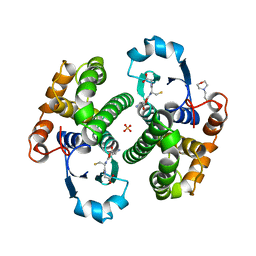 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
4PGT
 
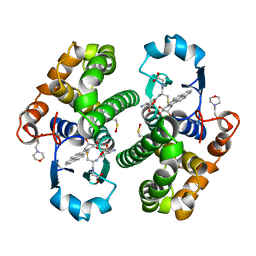 | | CRYSTAL STRUCTURE OF HGSTP1-1[V104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Blaszczyk, J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
2GSR
 
 | |
1B1U
 
 | |
5GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
6GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
1AQW
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1AQX
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH MEISENHEIMER COMPLEX | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1WUT
 
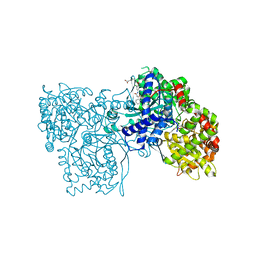 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | Descriptor: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
8G5C
 
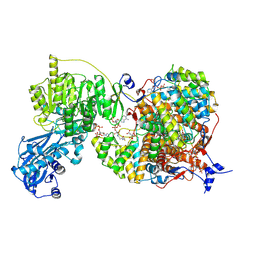 | |
6CV7
 
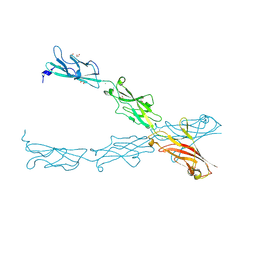 | |
1PGT
 
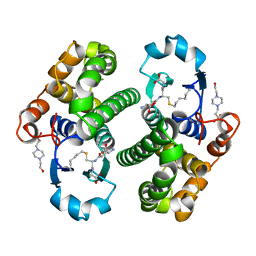 | |
6UUZ
 
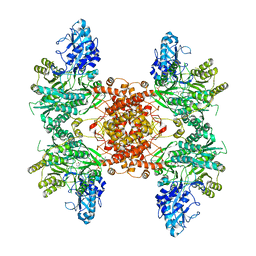 | |
5V5X
 
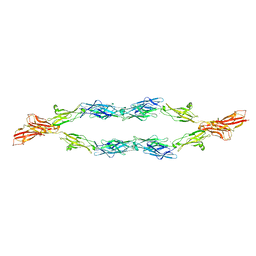 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GLR
 
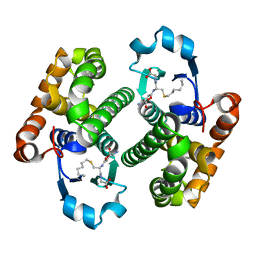 | |
7RGF
 
 | | Protocadherin gammaC4 EC1-4 crystal structure disrupted trans interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
2GSS
 
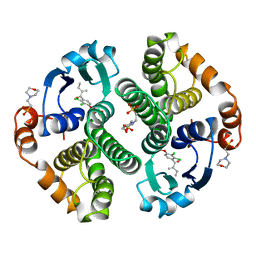 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
1GLQ
 
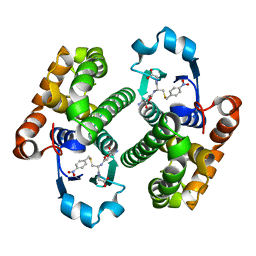 | |
5CEI
 
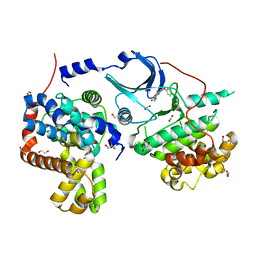 | | Crystal structure of CDK8:Cyclin C complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-iodophenoxy)-N-methylthieno[2,3-c]pyridine-2-carboxamide, Cyclin-C, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Koehler, M.F.T. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Development of a Potent, Specific CDK8 Kinase Inhibitor Which Phenocopies CDK8/19 Knockout Cells.
Acs Med.Chem.Lett., 7, 2016
|
|
5DEY
 
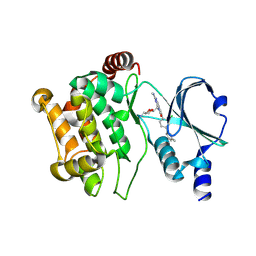 | | Crystal structure of PAK1 in complex with an inhibitor compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
5DZX
 
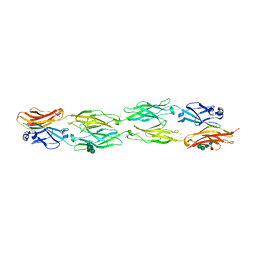 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5HVY
 
 | | CDK8/CYCC IN COMPLEX WITH COMPOUND 20 | | Descriptor: | CHLORIDE ION, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Bergeron, P, Koehler, M. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8.
Acs Med.Chem.Lett., 7, 2016
|
|
5DZW
 
 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5DZY
 
 | | Protocadherin beta 8 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Pcdhb8 protein, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
6TA8
 
 | |
