9H6E
 
 | |
9H3I
 
 | | trans-aconitate decarboxylase Tad1- wild type | | Descriptor: | Trans-aconitate decarboxylase 1 | | Authors: | Zheng, L, Bang, G. | | Deposit date: | 2024-10-16 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanistic and structural insights into the itaconate-producing trans -aconitate decarboxylase Tad1.
Pnas Nexus, 4, 2025
|
|
9H4G
 
 | |
9H4H
 
 | | trans-aconitate decarboxylase Tad1_S320A | | Descriptor: | Trans-aconitate decarboxylase 1 | | Authors: | Zheng, L, Bange, G. | | Deposit date: | 2024-10-18 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic and structural insights into the itaconate-producing trans -aconitate decarboxylase Tad1.
Pnas Nexus, 4, 2025
|
|
9H4E
 
 | |
9H1D
 
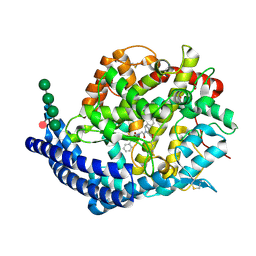 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD015 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-3-phenyl-2-sulfanyl-propanoyl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Mercapto-3-phenylpropanoyl Dipeptides as Dual Angiotensin-Converting Enzyme C-Domain-Selective/Neprilysin Inhibitors.
J.Med.Chem., 68, 2025
|
|
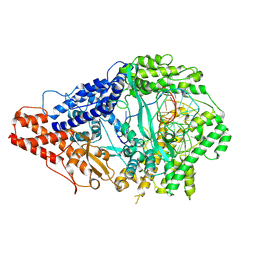
9FCV
 
 | |
9EOI
 
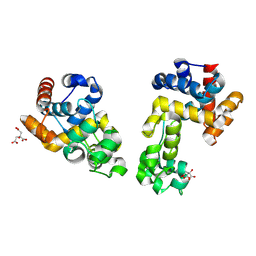 | |
9EOL
 
 | |
9F7V
 
 | |
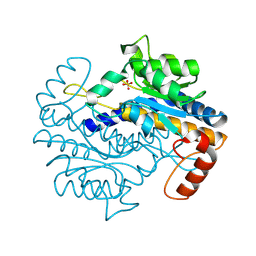
9F7O
 
 | |
9EWF
 
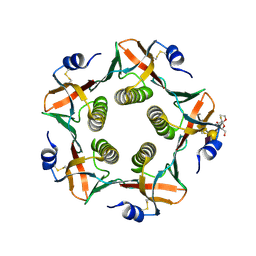 | | Cholera toxin B subunit in complex with fluorinated GM1 | | Descriptor: | (2R,3S,4S,5R,6R)-6-dodecoxy-5-fluoranyl-2-(hydroxymethyl)oxane-3,4-diol, 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, Cholera enterotoxin subunit B | | Authors: | Fan, J, Koehnke, J. | | Deposit date: | 2024-04-03 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Origin of Affinity in the GM1-Cholera Toxin Complex through Site-Selective Editing with Fluorine.
Acs Cent.Sci., 10, 2024
|
|
9EU4
 
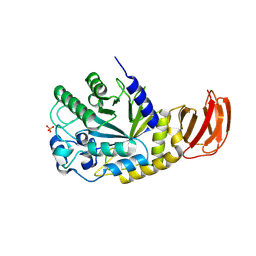 | | GH29A alpha-L-fucosidase | | Descriptor: | Exported alpha-L-fucosidase protein, SULFATE ION, beta-L-fucopyranose | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
9KC9
 
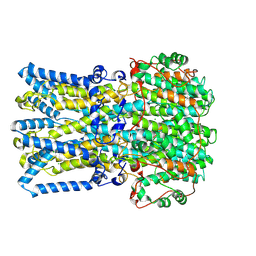 | | Cryo-EM structure of docked mouse bestrophin-1 in a partial open state | | Descriptor: | Bestrophin-1,Soluble cytochrome b562, CALCIUM ION, CHLORIDE ION | | Authors: | Lim, H.H, Kim, K.W, Ko, A. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of mouse bestrophin 1 channel in closed and partially open conformations.
Mol.Cells, 48, 2025
|
|
9JI6
 
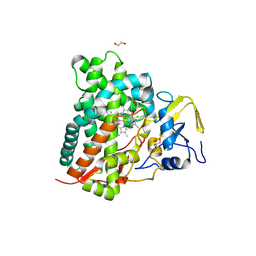 | | CYP105A1 R84A complexed with diclofenac (DIF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JIP
 
 | | CYP105A1 R84A complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-12 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JHW
 
 | | CYP105A1 complexed with diclofenac (DIF) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9JI1
 
 | | CYP105A1 complexed with flufenamic acid (FLF) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, ... | | Authors: | Mikami, B, Takita, T, Sakaki, T, Yasuda, K, Wada, M, Yasukawa, K. | | Deposit date: | 2024-09-11 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Function Analysis of Streptomyces griseolus CYP105A1 in the Metabolism of Nonsteroidal Anti-inflammatory Drugs.
Biochemistry, 64, 2025
|
|
9BA2
 
 | | Crystal structure of the binary complex of DCAF1 and WDR5 | | Descriptor: | DDB1- and CUL4-associated factor 1, IMIDAZOLE, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Srivastava, S, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9BD5
 
 | | Laccase from Bacillus licheniformis | | Descriptor: | COPPER (II) ION, SULFATE ION, Spore coat protein A | | Authors: | Habib, M.H, Smith, T.J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymerization potential of a bacterial CotA-laccase for beta-naphthol: enzyme structure and comprehensive polymer characterization.
Front Microbiol, 15, 2024
|
|
9B8J
 
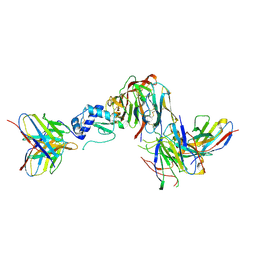 | | GP38-GnH-DS-Gc in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-36125 heavy chain, ADI-36125 light chain, ... | | Authors: | McFadden, E, McLellan, J.S. | | Deposit date: | 2024-03-30 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Engineering and structures of Crimean-Congo hemorrhagic fever virus glycoprotein complexes.
Cell, 188, 2025
|
|
9CS7
 
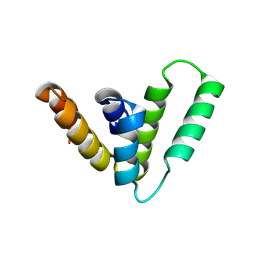 | | Structure of Azospirillum bacterial CARD crystal form 1 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, HIll, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
9CSF
 
 | |
9CS8
 
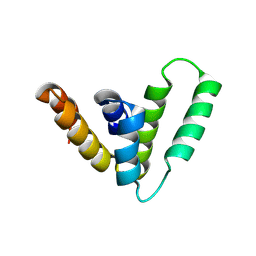 | | Structure of Azospirillum bacterial CARD crystal form 2 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, Hill, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
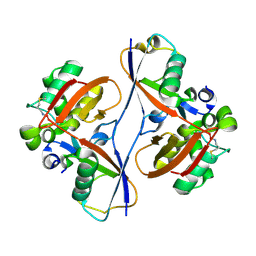
9CSH
 
 | |
