5OK6
 
 | | Ubiquitin specific protease 11 USP11 - peptide F complex | | Descriptor: | 1,2-ETHANEDIOL, ALA-GLU-GLY-GLU-PHE-TYR-LYS-LEU-LYS-ILE-ARG-THR-PRO-AAR, GLYCEROL, ... | | Authors: | Spiliotopoulos, A, Dreveny, I. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11.
J.Biol.Chem., 294, 2019
|
|
5PGU
 
 | |
5PGX
 
 | |
5N2P
 
 | | Sulfolobus solfataricus Tryptophan Synthase A | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Tryptophan synthase alpha chain | | Authors: | Fleming, J, Mayans, O. | | Deposit date: | 2017-02-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
J.Mol.Biol., 430, 2018
|
|
5PGY
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGZ
 
 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
6PI7
 
 | |
7SR4
 
 | |
7ST3
 
 | |
7STG
 
 | |
7SR0
 
 | | Single chain trimer HLA-A*02:01 (H98L, Y108C) with HPV.16 E7 peptide YMLDLQPET | | Descriptor: | PHOSPHATE ION, Protein E7 peptide,Beta-2-microglobulin,MHC class I antigen chimera, VHH | | Authors: | Finton, K.A.K, Rupert, P.B. | | Deposit date: | 2021-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Effects of HLA single chain trimer design on peptide presentation and stability.
Front Immunol, 14, 2023
|
|
7SR3
 
 | |
7SR5
 
 | |
7SSH
 
 | |
7T3E
 
 | | Structure of the sialic acid bound Tripartite ATP-independent Periplasmic (TRAP) periplasmic component SiaP from Photobacterium profundum | | Descriptor: | N-acetyl-beta-neuraminic acid, SULFATE ION, TRAP-type C4-dicarboxylate transport system, ... | | Authors: | Davies, J.S, Currie, M.J, North, R.A, Dobson, R.C.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
7T1D
 
 | | Human SIRT2 in complex with small molecule 359 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2,4-dimethyl-1H-imidazol-1-yl)-2-(5-{[4-(1H-pyrazol-1-yl)phenyl]methyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Kulp, J.L, Remiszewski, S, Todd, M, Chiang, L.W. | | Deposit date: | 2021-12-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric inhibitor of sirtuin 2 deacetylase activity exhibits broad-spectrum antiviral activity.
J.Clin.Invest., 133, 2023
|
|
7SQB
 
 | | PPAR gamma LBD bound to Inverse Agonist SR10221 | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
7SQA
 
 | | PPAR gamma LBD bound to SR10221 and SMRT corepressor motif | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
7TLY
 
 | |
7TLZ
 
 | |
7TM0
 
 | |
7U0D
 
 | |
7TTP
 
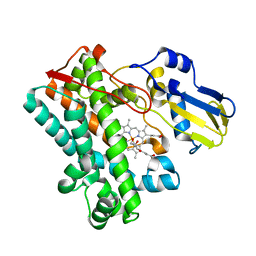 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTA
 
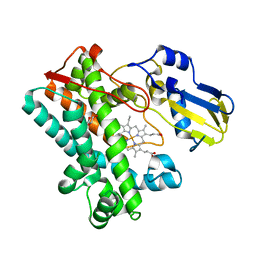 | | P450 (OxyA) from kistamicin biosynthesis, mixed heme conformation, attenuated beam | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
7TTQ
 
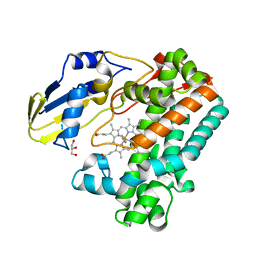 | | P450 (OxyA) from kistamicin biosynthesis, imidazole complex | | Descriptor: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
