4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6O30
 
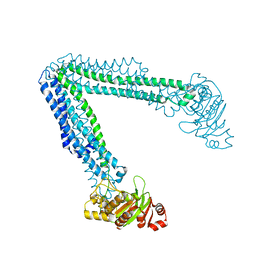 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
3QPS
 
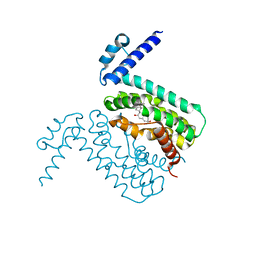 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
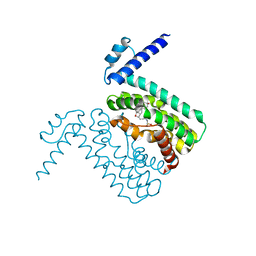 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
5XPV
 
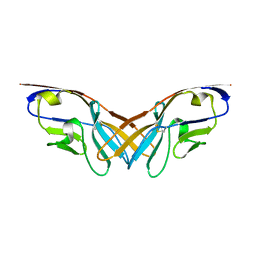 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
6A96
 
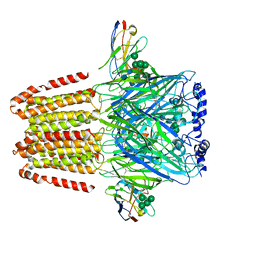 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
6INB
 
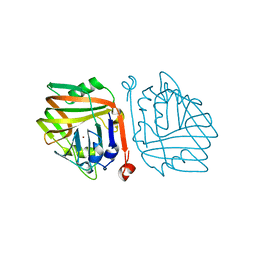 | |
6INC
 
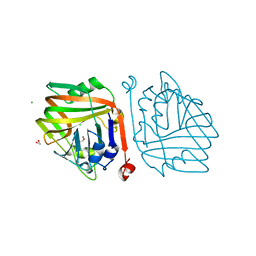 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
2MHI
 
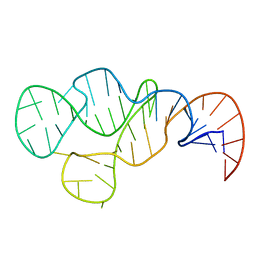 | |
8I8A
 
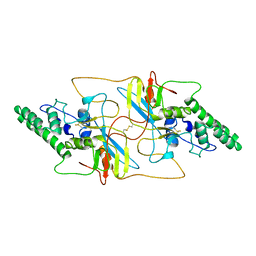 | |
8I8C
 
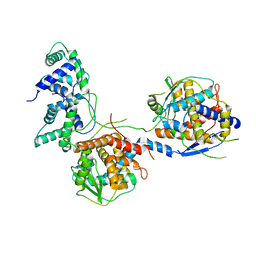 | |
8I8B
 
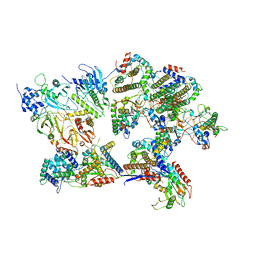 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
6JC4
 
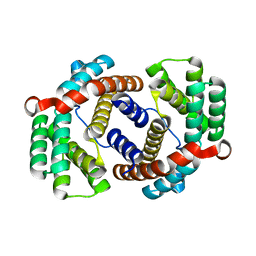 | |
2QCO
 
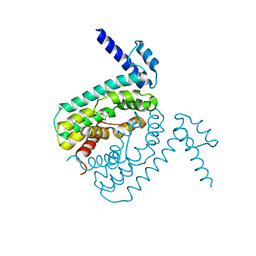 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
8IJR
 
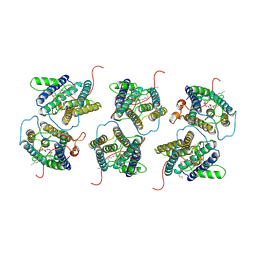 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5WCH
 
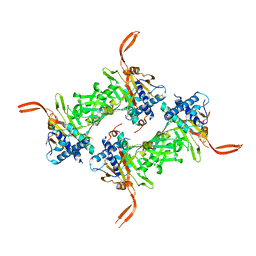 | | Crystal structure of the catalytic domain of human USP9X | | Descriptor: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5X0R
 
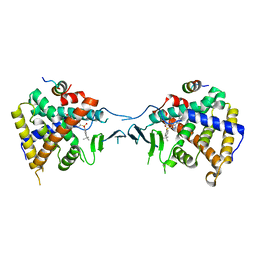 | | Crystal Structure of PXR LBD Complexed with SJB7 | | Descriptor: | 4-[(4-tert-butylphenyl)sulfonyl]-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Lv, L, Lin, W, Chai, S.C, Zhang, Q, Chen, T. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | SPA70 is a potent antagonist of human pregnane X receptor.
Nat Commun, 8, 2017
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
6BL6
 
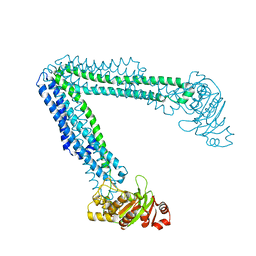 | | Crystallization of lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Stanfield, R.L, Zhang, Q, Wilson, I.A, Lee, S.C. | | Deposit date: | 2017-11-09 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
5VN7
 
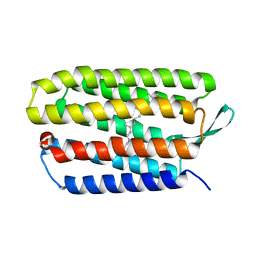 | | Structure of bacteriorhodopsin from crystals grown at 20 deg Celcius using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
5Z2V
 
 | | Crystal structure of RecR from Pseudomonas aeruginosa PAO1 | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6JYV
 
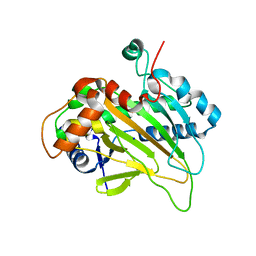 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | Descriptor: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | Authors: | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
