6V1L
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1B
 
 | | Crystal structure of the bromodomain of human BRD9 bound to I-BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1F
 
 | | Crystal structure of the bromodomain of human BRD7 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6VGL
 
 | | JAK2 JH1 in complex with ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
3CRZ
 
 | | Ferredoxin-NADP Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP+ reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Han, H, Schonbrunn, E. | | Deposit date: | 2008-04-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic and solution state nuclear magnetic resonance spectroscopic investigations of NADP+ binding to ferredoxin NADP reductase from Pseudomonas aeruginosa
Biochemistry, 47, 2008
|
|
4O70
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with DINACICLIB | | Descriptor: | 1,2-ETHANEDIOL, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-251527 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-2-(2-methoxyphenoxy)pyrimidine, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7A
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-409514 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-chloro-4-hydroxyphenyl)amino]-4-(3-chlorophenyl)-1H-pyrrole-2,5-dione, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7C
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-614067-R | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5Z)-5-(1-nitroso-2,3-dihydro-5H-inden-5-ylidene)-2-(piperidin-4-yl)-3,5-dihydro-4H-imidazol-4-ylidene]-1,4-dihydropyridine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7E
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-610251-B | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-phenyl-4-(pyridin-4-yl)-1H-imidazol-5-yl]phenol, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O74
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with BI 2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O7B
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-284847-BT | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-dimethylphenoxy)-4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]pyrimidine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O78
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with GW612286X | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O76
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with TG101209 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O77
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB 202190 | | Descriptor: | 4-[4-(4-fluorophenyl)-5-(pyridin-4-yl)-1H-imidazol-2-yl]phenol, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O71
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FLAVOPIRIDOL | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O75
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FOSTAMATINIB | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [6-({5-fluoro-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}amino)-2,2-dimethyl-3-oxo-2,3-dihydro-4H-pyrido[3,2-b][1,4]oxazin-4-yl]methyl dihydrogen phosphate | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
2PQC
 
 | | CP4 EPSPS liganded with (R)-phosphonate tetrahedral reaction intermediate analog | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQB
 
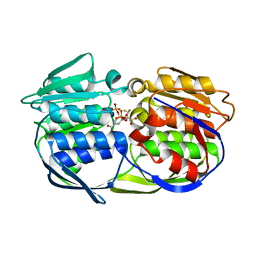 | | CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQD
 
 | | A100G CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral reaction intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2QDX
 
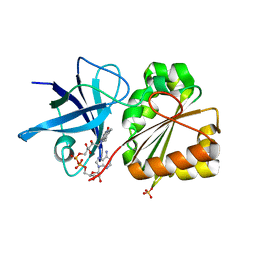 | | P.Aeruginosa Fpr with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase, SULFATE ION | | Authors: | Han, H, Schonbrunn, E. | | Deposit date: | 2007-06-21 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP(+) Reductase and Not Ferredoxin Is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions
Biochemistry, 46, 2007
|
|
4ERW
 
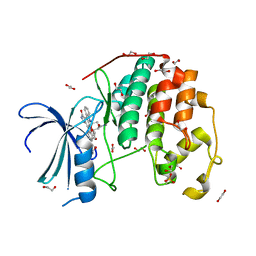 | | CDK2 in complex with staurosporine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, STAUROSPORINE | | Authors: | Alam, R, Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4DEB
 
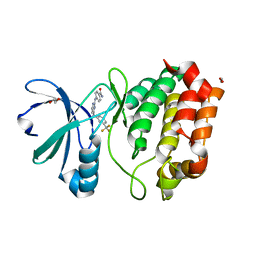 | | Aurora A in complex with RK2-17-01 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[3-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzamide, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
