6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | Descriptor: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | Authors: | Zhang, X, Ma, J, Chen, L. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
8WRQ
 
 | |
8K52
 
 | | Cryo-EM structure of chitin synthase | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | Authors: | Zhang, X, Niu, S, Li, P, Bi, Y. | | Deposit date: | 2023-07-20 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
7CTH
 
 | | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, Single Chain Variable Fragment | | Authors: | Zhang, X, Sharma, A, Duquerroy, S, Zhou, Z.H, Rey, F.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
8Z0O
 
 | | Cryo-EM structure of chitin synthase | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | Authors: | Zhang, X, Niu, S, Li, P, Bi, Y. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
7DY9
 
 | | Thermotoga maritima ferritin mutant-FLAL | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DYB
 
 | | Thermotoga maritima ferritin mutant-FLAL-L | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DV8
 
 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FFO
 
 | | Cryo-EM structure of VEEV VLP at the 5-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFQ
 
 | | Cryo-EM structure of VEEV VLP at the 2-fold axes | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFE
 
 | | Cryo-EM structure of VEEV VLP | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFF
 
 | | Structure of Venezuelan equine encephalitis virus with the receptor LDLRAD3 | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFL
 
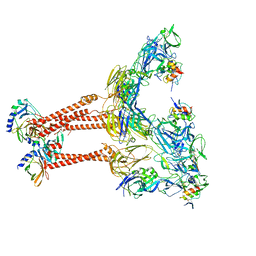 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7FFN
 
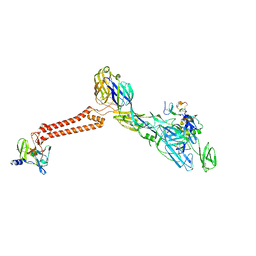 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 5-fold axes | | Descriptor: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | Authors: | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | Deposit date: | 2021-07-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7VH2
 
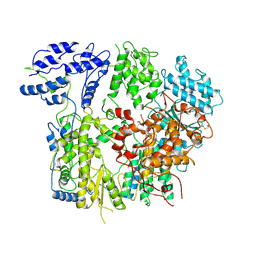 | |
7VGQ
 
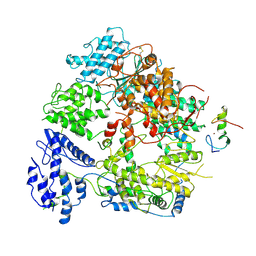 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-18 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH1
 
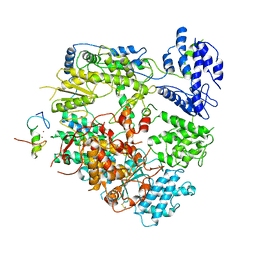 | | Cryo-EM structure of Machupo virus dimeric L-Z complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-20 | | Release date: | 2021-09-29 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7V66
 
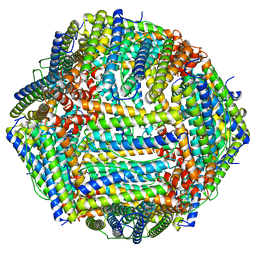 | | Structure of Apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, X, Wu, C, Shi, H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
7X5B
 
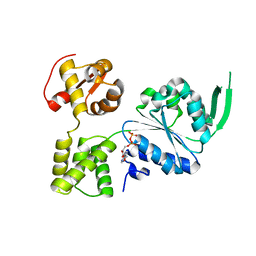 | | Crystal structure of RuvB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5A
 
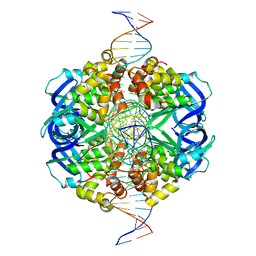 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9LNP
 
 | | the hUNG bound to DNA product embedding uridine ribonucleotide | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*T*(RP5)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
9LNQ
 
 | | The hUNG bound to DNA product embedding 4primer-OCH3-dU | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(KBC)P*AP*TP*CP*TP*T)-3'), SODIUM ION, ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
