6XBK
 
 | | Structure of human SMO-G111C/I496C complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XBM
 
 | | Structure of human SMO-Gi complex with 24(S),25-EC | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XBL
 
 | | Structure of human SMO-Gi complex with SAG | | Descriptor: | 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
1VM2
 
 | |
1VM4
 
 | |
1VM5
 
 | |
1VM3
 
 | |
8TZS
 
 | | Structure of human WLS | | Descriptor: | Protein wntless homolog | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZP
 
 | | Structure of human Wnt7a bound to WLS and RECK | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PALMITOLEIC ACID, Protein Wnt-7a, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZR
 
 | | Structure of human Wnt3a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calreticulin, PALMITOLEIC ACID, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZO
 
 | | Structure of human Wnt7a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, Calreticulin, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
6LTU
 
 | | Crystal structure of Cas12i2 ternary complex with double Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
7A8P
 
 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
5C4W
 
 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
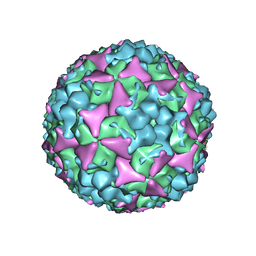 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
6OEU
 
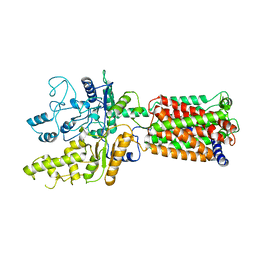 | | Structure of human Patched1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Qi, X, Li, X, Wang, J. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
5DPM
 
 | |
6OEV
 
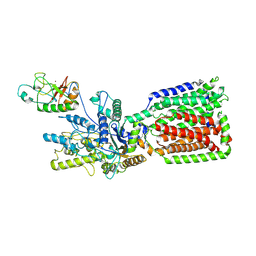 | | Structure of human Patched1 in complex with native Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-17 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
6OT0
 
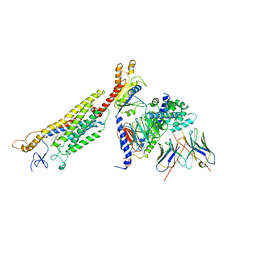 | | Structure of human Smoothened-Gi complex | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Fab heavy chain, Fab light chain, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi.
Nature, 571, 2019
|
|
5C8C
 
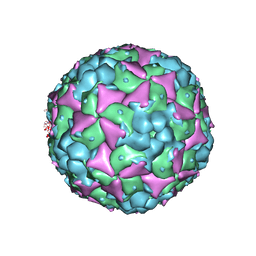 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
9N0Y
 
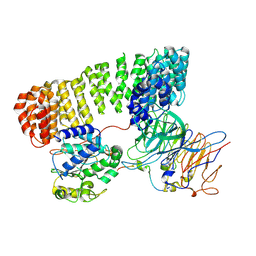 | | PP2A-B55 Holoenzyme with Eya3 | | Descriptor: | Protein phosphatase EYA3, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Shi, S, Li, X, Alderman, C, Zhao, R. | | Deposit date: | 2025-01-24 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cryo-EM structures reveal the PP2A-B55 alpha and Eya3 interaction that can be disrupted by a peptide inhibitor.
J.Biol.Chem., 301, 2025
|
|
9N0Z
 
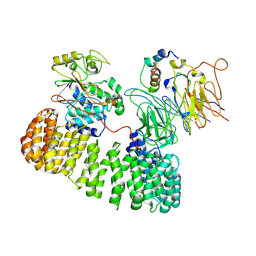 | | PP2A-B55 Holoenzyme with B55i | | Descriptor: | B55i inhibitor peptide, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Shi, S, Li, X, Alderman, C, Zhao, R. | | Deposit date: | 2025-01-24 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal the PP2A-B55 alpha and Eya3 interaction that can be disrupted by a peptide inhibitor.
J.Biol.Chem., 301, 2025
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
3VBS
 
 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
