1TE6
 
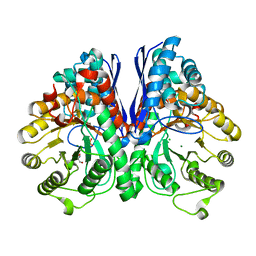 | | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Gamma enolase, ... | | Authors: | Chai, G, Brewer, J, Lovelace, L, Aoki, T, Minor, W, Lebioda, L. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Purification and the 1.8 A Resolution Crystal Structure of Human Neuron Specific Enolase
J.Mol.Biol., 341, 2004
|
|
5NLS
 
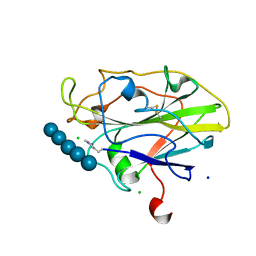 | | Auxiliary activity 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates.
Nat Commun, 8, 2017
|
|
3NMW
 
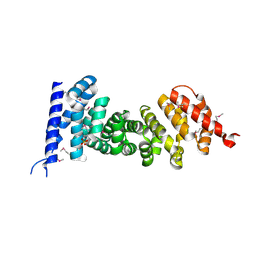 | | Crystal structure of armadillo repeats domain of APC | | Descriptor: | APC variant protein, SULFATE ION | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3DLQ
 
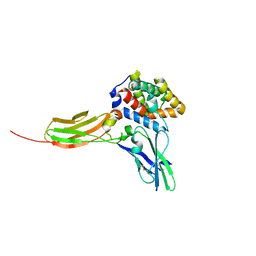 | | Crystal structure of the IL-22/IL-22R1 complex | | Descriptor: | Interleukin-22, Interleukin-22 receptor subunit alpha-1 | | Authors: | Bleicher, L, de Moura, P.R, Watanabe, L, Colau, D, Dumoutier, L, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2008-06-28 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism
Febs Lett., 582, 2008
|
|
6NDO
 
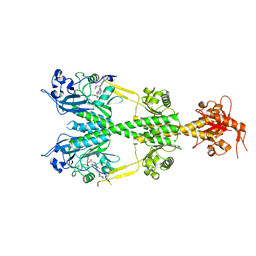 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193N from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
3OF0
 
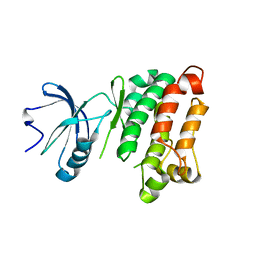 | |
1A3J
 
 | | X-RAY CRYSTALLOGRAPHIC DETERMINATION OF A COLLAGEN-LIKE PEPTIDE WITH THE REPEATING SEQUENCE (PRO-PRO-GLY) | | Descriptor: | COLLAGEN-LIKE PEPTIDE | | Authors: | Kramer, R.Z, Vitagliano, L, Bella, J, Berisio, R, Mazzarella, L, Brodsky, B, Zagari, A, Berman, H.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly).
J.Mol.Biol., 280, 1998
|
|
8OTL
 
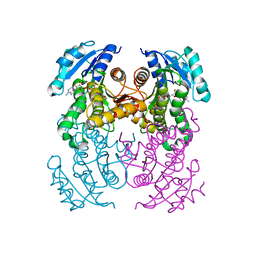 | | structure of InhA from Mycobacterium tuberculosis in complex with 5-(((4-(2-hydroxyphenoxy)benzyl)(octyl)amino)methyl)-2-phenoxyphenol | | Descriptor: | 1,2-ETHANEDIOL, 5-[[octyl-[[4-(2-oxidanylphenoxy)phenyl]methyl]amino]methyl]-2-phenoxy-phenol, ACETATE ION, ... | | Authors: | Tamhaev, R, Maveyraud, L, Chebaiki, M, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Exploring the plasticity of the InhA substrate-binding site using new diaryl ether inhibitors.
Bioorg.Chem., 143, 2023
|
|
5CXD
 
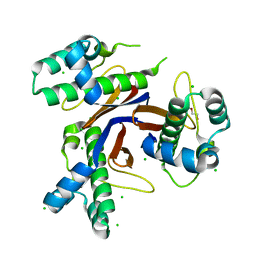 | | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Holo-[acyl-carrier-protein] synthase, ... | | Authors: | Halavaty, A.S, Minasov, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
To Be Published
|
|
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
5NPC
 
 | | Crystal Structure of D412N nucleophile mutant cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with unreacted alpha Cyclophellitol Cyclosulfate probe ME647 | | Descriptor: | (3~{a}~{R},4~{R},5~{S},6~{R},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5NLT
 
 | | CvAA9A | | Descriptor: | COPPER (II) ION, CvAA9A, SULFATE ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tandrup, T, Schnorr, K, Tovborg, M, Lo Leggio, L. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates.
Nat Commun, 8, 2017
|
|
8H95
 
 | | Structure of mouse SCMC bound with full-length FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, L, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5LWP
 
 | | Discovery of phenoxyindazoles and phenylthioindazoles as RORg inverse agonists | | Descriptor: | 4-[3-[2-chloranyl-6-(trifluoromethyl)phenoxy]-5-(dimethylcarbamoyl)indazol-1-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Ouvry, G, Bouix-Peter, C, Ciesielski, F, Chantalat, L, Christin, O, Comino, C, Duvert, D, Feret, C, Harris, C.S, Luzy, A.-P, Musicki, B, Orfila, D, Pascau, J, Parnet, V, Perrin, A, Pierre, R, Raffin, C, Rival, Y, Taquet, N, Thoreau, E, Hennequin, L.F. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of phenoxyindazoles and phenylthioindazoles as ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
2B8W
 
 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
1V04
 
 | | serum paraoxonase by directed evolution | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SERUM PARAOXONASE/ARYLESTERASE 1 | | Authors: | Harel, M, Aharoni, A, Gaidukov, L, Brumshtein, B, Khersonsky, O, Yagur, S, Meged, R, Dvir, H, Ravelli, R.B.G, McCarthy, A, Toker, L, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-23 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Evolution of the Serum Paraoxonase Family of Detoxifying and Anti-Atherosclerotic Enzymes
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UY7
 
 | | Human Hsp90-alpha with 9-Butyl-8-(4-methoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(4-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
8BB6
 
 | |
1UYC
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
5NHH
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHO
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{S})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHV
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NGU
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
