5ZUO
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5FXL
 
 | |
2BU3
 
 | | Acyl-enzyme intermediate between Alr0975 and glutathione at pH 3.4 | | Descriptor: | ALR0975 PROTEIN, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivares, D, Arnoux, P, Pignol, D. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Papain-Like Enzyme at Work: Native and Acyl- Enzyme Intermediate Structures in Phytochelatin Synthesis.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1BWO
 
 | |
1URF
 
 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
1UQV
 
 | | SAM domain from Ste50p | | Descriptor: | STE50 PROTEIN | | Authors: | Grimshaw, S.J, Mott, H.R, Stott, K.M, Nielsen, P.R, Evetts, K.A, Hopkins, L.J, Nietlispach, D, Owen, D. | | Deposit date: | 2003-10-20 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sterile {Alpha} Motif (Sam) Domain of the Saccharomyces Cerevisiae Mitogen-Activated Protein Kinase Pathway-Modulating Protein Ste50 and Analysis of its Interaction with the Ste11 Sam
J.Biol.Chem., 279, 2004
|
|
2YOA
 
 | | Synaptotagmin-1 C2B domain with phosphoserine | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, SYNAPTOTAGMIN-1, ... | | Authors: | Honigmann, A, van den Bogaart, G, Iraheta, E, Risselada, H.J, Milovanovic, D, Mueller, V, Muellar, S, Diederichsen, U, Fasshauer, D, Grubmuller, H, Hell, S.W, Eggeling, C, Kuhnel, K, Jahn, R. | | Deposit date: | 2012-10-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Clusters Act as Molecular Beacons for Vesicle Recruitment
Nat.Struct.Mol.Biol., 20, 2013
|
|
1UUH
 
 | | Hyaluronan binding domain of human CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Teriete, P, Banerji, S, Noble, M, Blundell, C, Wright, A, Pickford, A, Lowe, E, Mahoney, D, Tammi, M, Kahmann, J, Campbell, I, Day, A, Jackson, D. | | Deposit date: | 2003-12-19 | | Release date: | 2004-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Regulatory Hyaluronan-Binding Domain in the Inflammatory Leukocyte Homing Receptor Cd44
Mol.Cell, 13, 2004
|
|
5BTR
 
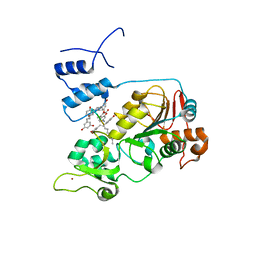 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
4PV2
 
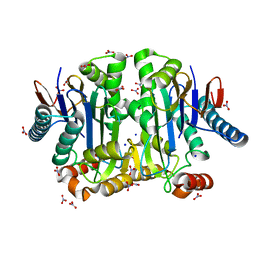 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3CBP
 
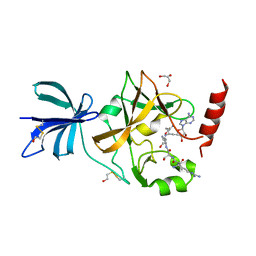 | | Set7/9-ER-Sinefungin complex | | Descriptor: | BETA-MERCAPTOETHANOL, Estrogen receptor, GLYCEROL, ... | | Authors: | Cheng, X, Jia, D. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase.
Mol.Cell, 30, 2008
|
|
5ARG
 
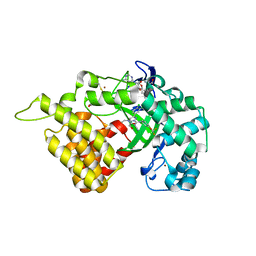 | | SMYD2 in complex with SGC probe BAY-598 | | Descriptor: | GLYCEROL, N-LYSINE METHYLTRANSFERASE SMYD2, N-[1-(N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL)-3-(3,4-DICHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
1Z1M
 
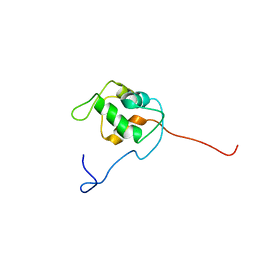 | | NMR structure of unliganded MDM2 | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Uhrinova, S, Uhrin, D, Powers, H, Watt, K, Zheleva, D, Fischer, P, McInnes, C, Barlow, P.N. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Free MDM2 N-terminal Domain Reveals Conformational Adjustments that Accompany p53-binding
J.Mol.Biol., 350, 2005
|
|
5G5C
 
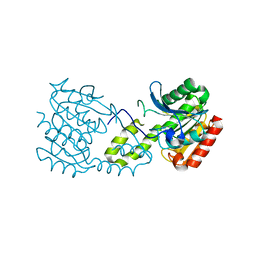 | |
1XI9
 
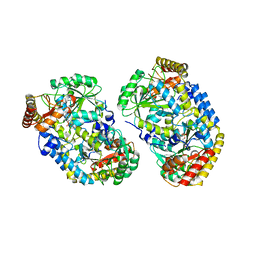 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
3AGL
 
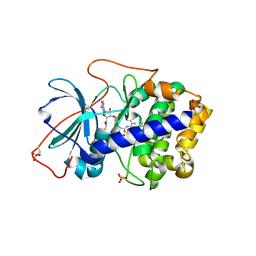 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1039 | | Descriptor: | (10R,20R,23R)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamido propyl)-10-methyl-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
5G59
 
 | |
1RT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH MKC-442 | | Descriptor: | 6-BENZYL-1-ETHOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
2EXI
 
 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
1XN9
 
 | | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11 | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methanosarcina mazei Protein RPS24E: The Northeast Structural Genomics Consortium Target MaR11
To be Published
|
|
1XX7
 
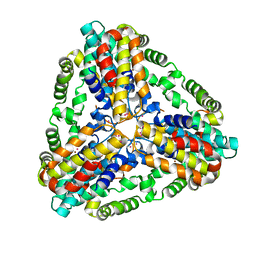 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001 | | Descriptor: | NICKEL (II) ION, UNKNOWN ATOM OR ION, oxetanocin-like protein | | Authors: | Chen, L, Tempel, W, Habel, J, Zhou, W, Nguyen, D, Chang, S.-H, Lee, D, Kelley, L.-L.C, Dillard, B.D, Liu, Z.-J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001
To be published
|
|
1XAV
 
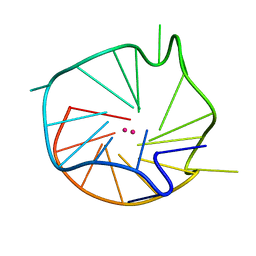 | | Major G-quadruplex structure formed in human c-MYC promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3', POTASSIUM ION | | Authors: | Ambrus, A, Chen, D, Dai, J, Jones, R.A, Yang, D. | | Deposit date: | 2004-08-26 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the biologically relevant G-Quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization.
Biochemistry, 44, 2005
|
|
2FLD
 
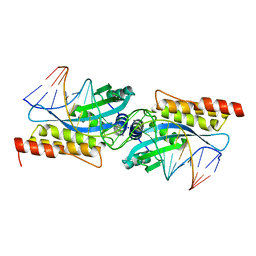 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
2BTW
 
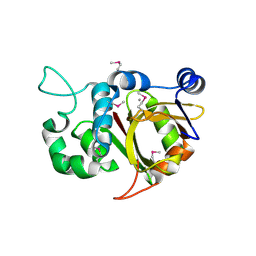 | | Crystal structure of Alr0975 | | Descriptor: | ALR0975 PROTEIN, CALCIUM ION | | Authors: | Vivares, D, Arnoux, P, Pignol, D. | | Deposit date: | 2005-06-07 | | Release date: | 2005-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Papain-Like Enzyme at Work: Native and Acyl- Enzyme Intermediate Structures in Phytochelatin Synthesis.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
