3L4U
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with de-O-sulfonated kotalanol | | Descriptor: | (2R,3S,4S)-1-[(2S,3S,4R,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl]-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
3L4Y
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8II | | Descriptor: | (1S,2R,3S,4R)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
8E4H
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E3R
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E3K
 
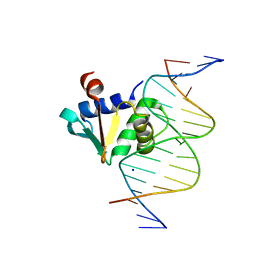 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E5Y
 
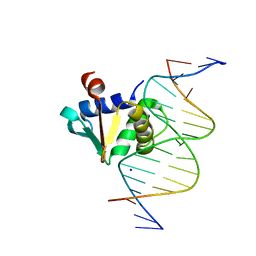 | |
8EBH
 
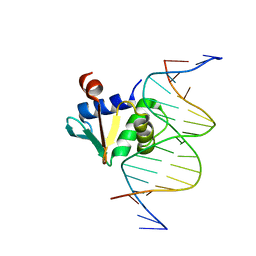 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-31 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
3G9L
 
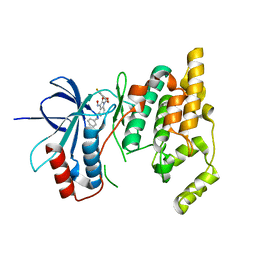 | |
5WJN
 
 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
3G9N
 
 | |
7DNF
 
 | | DARPin 63_B7 in complex with V3-IY (MN) crown mimetic | | Descriptor: | DARPin 63_B7, SULFATE ION, V3-IY (MN) crown mimetic peptide | | Authors: | Wu, Y, Plueckthun, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7DNG
 
 | |
7DNE
 
 | |
8DR0
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQX
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
1ML4
 
 | |
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR6
 
 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
3L4X
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8 | | Descriptor: | (1S,2R,3S,4S)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
3L4T
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with BJ2661 | | Descriptor: | (1R,2S)-1-[(1S)-1,2-dihydroxyethyl]-3-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-2-hydroxypropyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltase-glucoamylase, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
8UOS
 
 | |
8UPB
 
 | |
8UPA
 
 | | Structure of gp130 in complex with a de novo designed IL-6 mimetic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, De novo designed IL-6 mimetic, Interleukin-6 receptor subunit beta, ... | | Authors: | Borowska, M.T, Jude, K.M, Garcia, K.C. | | Deposit date: | 2023-10-22 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo design of miniprotein antagonists of cytokine storm inducers.
Nat Commun, 15, 2024
|
|
7XWX
 
 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
