9OXQ
 
 | |
9LVR
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, 6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-1-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9M5M
 
 | | Crystal structure of Plasmodium vivax aspartyl-tRNA synthetase (PvDRS) | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, aspartate--tRNA ligase | | Authors: | Sharma, V.K, Manickam, Y, Bagale, S, Pradeepkumar, P.I, Sharma, A. | | Deposit date: | 2025-03-06 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Natural product-mediated reaction hijacking mechanism validates Plasmodium aspartyl-tRNA synthetase as an antimalarial drug target
To Be Published
|
|
9NLP
 
 | | HIV-1 Reverse Transcriptase with New Non-Nucleoside Reverse Transcriptase Inhibitor 12126065 | | Descriptor: | 4-({5-amino-1-[6-(2-cyanoethyl)naphthalene-1-sulfonyl]-1H-1,2,4-triazol-3-yl}amino)-2-chlorobenzonitrile, Reverse transcriptase p51 subunit, Reverse transcriptase p66 subunit | | Authors: | Young, M.A, Lane, T.R, Raman, R, Nelson, J.A.E, Riabova, O, Kazakova, E, Monakhova, N, Tsedilin, A, Rees, S.D, Quinnell, D, Chang, G, Ekins, S. | | Deposit date: | 2025-03-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM Structure of HIV-1 Reverse Transcriptase with N -Phenyl-1-(phenylsulfonyl)-1 H -1,2,4-triazol-3-amine: A New HIV-1 Non-nucleoside Inhibitor.
Acs Infect Dis., 11, 2025
|
|
9QGD
 
 | | Crystal structure of an NADH-accepting ene reductase variant NostocER1-L1,5 mutant Q350K | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, All1865 protein, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Maslakova, A, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Rational Introduction of Electrostatic Interactions at Crystal Contacts to Enhance Protein Crystallization of an Ene Reductase.
Biomolecules, 15, 2025
|
|
9NQA
 
 | | [0,9,10-2] Shifted tensegrity triangle with an (arm,center,arm) distribution of (0,9,10) base pairs and 2 nt sticky ends containing a semi-junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*TP*GP*CP*GP*GP*TP*GP*T)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*TP*GP*GP*AP*CP*AP*CP*CP*GP*CP*AP*TP*C)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (5.47 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9QFV
 
 | | Human Tryptase beta-2 (hTPSB2) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Porta, A, Manelfi, C, Talarico, C, Beccari, A.R, Brindisi, M, Summa, V, Iaconis, D, Gobbi, M, Beeg, M. | | Deposit date: | 2025-03-12 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Integrating Surface Plasmon Resonance and Docking Analysis for Mechanistic Insights of Tryptase Inhibitors.
Molecules, 30, 2025
|
|
9QGE
 
 | | Crystal structure of an NADH-accepting ene reductase variant NostocER1-L1,5 mutant D352K | | Descriptor: | 1,2-ETHANEDIOL, All1865 protein, CHLORIDE ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Maslakova, A, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | Rational Introduction of Electrostatic Interactions at Crystal Contacts to Enhance Protein Crystallization of an Ene Reductase.
Biomolecules, 15, 2025
|
|
9OWG
 
 | | Crystal Structure of the Immunity Protein (52 domain-containing protein) from Pseudomonas aeruginosa PAO1. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBONATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Stogios, P.J, Savchenko, A, Satchell, K.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-02 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Immunity Protein (52 domain-containing protein) from Pseudomonas aeruginosa PAO1.
To Be Published
|
|
9LVV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 17 (S-892216) | | Descriptor: | 1-(2-azanylideneethyl)-6-[6,6-bis(fluoranyl)-2-azaspiro[3.3]heptan-2-yl]-5-(3-chloranyl-4-fluoranyl-phenyl)-3-(5-chloranylpyridin-3-yl)pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9QEK
 
 | | Structure of Human ROS1 Kinase Domain Harboring the G2032R Solvent-front Mutation in Complex with Zidesamtinib (NVL-520) | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase ROS, Zidesamtinib | | Authors: | Tangpeerachaikul, A, Mente, S, Magrino, J, Gu, F, Horan, J.C, Pelish, H.E. | | Deposit date: | 2025-03-10 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Zidesamtinib Selective Targeting of Diverse ROS1 Drug-Resistant Mutations.
Mol.Cancer Ther., 2025
|
|
9UGE
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution | | Descriptor: | D-MALATE, OXALIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Barik, D, Ahmad, N, Maurya, A, Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution
To Be Published
|
|
6WS6
 
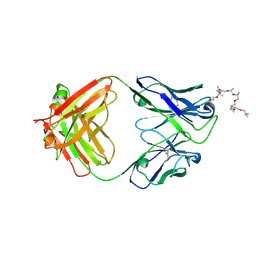 | | Structural and functional analysis of a potent sarbecovirus neutralizing antibody | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), S309 antigen-binding (Fab) fragment, heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, Marco, A.D, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Telenti, A, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
7NP4
 
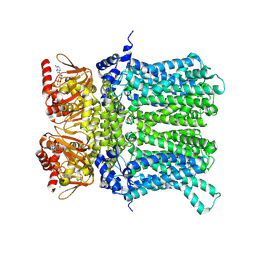 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
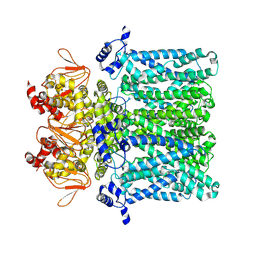 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
8X5U
 
 | | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase C-terminal 16 amino acid deletion mutant | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Uehara, Y, Matsumoto, A, Nakazawa, T, Fukuta, A, Ando, K, Oka, N, Uchiumi, T, Ito, K. | | Deposit date: | 2023-11-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding mode between peptidyl-tRNA hydrolase and the peptidyl-A76 moiety of the substrate.
J.Biol.Chem., 301, 2025
|
|
8X5T
 
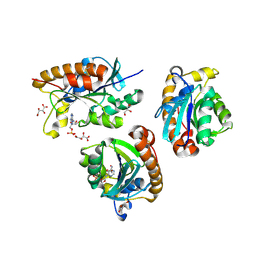 | | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase in complex with adenosine 5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Uehara, Y, Matsumoto, A, Nakazawa, T, Fukuta, A, Ando, K, Oka, N, Uchiumi, T, Ito, K. | | Deposit date: | 2023-11-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode between peptidyl-tRNA hydrolase and the peptidyl-A76 moiety of the substrate.
J.Biol.Chem., 301, 2025
|
|
8UDF
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor DEL_7 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-1-(4-fluorophenyl)-4-(methylamino)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor DEL_2 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-(methylamino)-4-oxo-1-phenylbutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UEF
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 32 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-{(2S)-1-(4-fluorophenyl)-4-[(4-methoxybutyl)amino]-4-oxobutan-2-yl}-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Choi, K, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-10-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDO
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 15 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-{[5-(dimethylamino)pentyl]amino}-1-(4-fluorophenyl)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDQ
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 1 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-{[2-(dimethylamino)ethyl]amino}-1-(4-fluorophenyl)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5, SODIUM ION | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDW
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 2 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-{[3-(dimethylamino)propyl]amino}-1-(4-fluorophenyl)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-29 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UDP
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 14 | | Descriptor: | 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-4-{[4-(dimethylamino)butyl]amino}-1-(4-fluorophenyl)-4-oxobutan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5 | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-28 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
8UEB
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-1-(4-fluorophenyl)-4-oxo-4-{[3-(pyridin-4-yl)propyl]amino}butan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Choi, K, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-30 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
