4B5Y
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise-GL (K206A mutant) in space group C222(1) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Lelimousin, M, Oost, K, Noirclerc-Savoye, M, Gadella, T.W.J, Goedhart, J, Royant, A. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of the H148G Mutation on Fluorescence Properties of Cyan Fluorescent Proteins
To be Published
|
|
8BJW
 
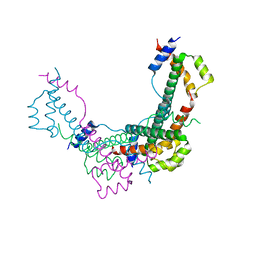 | |
8B0Q
 
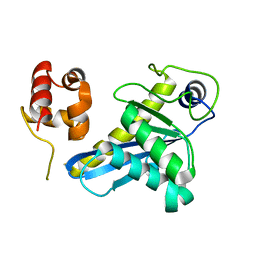 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
6GHV
 
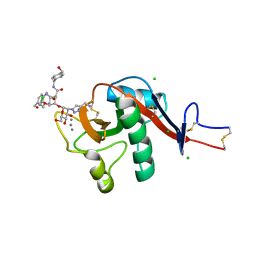 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
4NCG
 
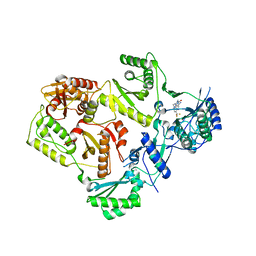 | | Discovery of Doravirine, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of MK-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2XR6
 
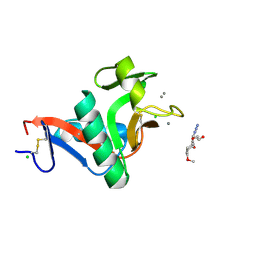 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
6HKT
 
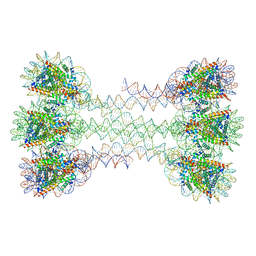 | | Structure of an H1-bound 6-nucleosome array | | Descriptor: | DNA (1122-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Garcia-Saez, I, Dimitrov, S, Petosa, C. | | Deposit date: | 2018-09-08 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (9.7 Å) | | Cite: | Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation.
Mol. Cell, 72, 2018
|
|
6WOJ
 
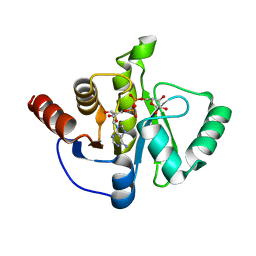 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
7KUW
 
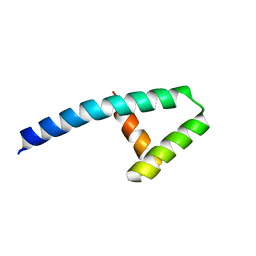 | |
8C3W
 
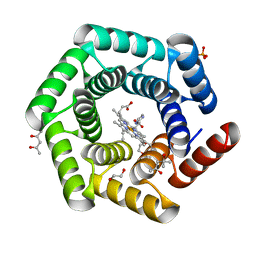 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
9CKV
 
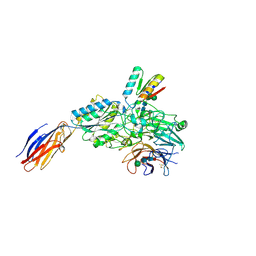 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | De Novo Design of Integrin alpha 5 beta 1 Modulating Proteins for Regenerative Medicine.
Biorxiv, 2024
|
|
3IHC
 
 | |
3IIG
 
 | |
3ILB
 
 | |
3IHD
 
 | |
3IHF
 
 | |
3IIH
 
 | |
3IHE
 
 | |
3ILC
 
 | |
7QVB
 
 | |
5IXE
 
 | | 1.75A RESOLUTION STRUCTURE OF 5-Fluoroindole BOUND BETA-GLYCOSIDASE (W33G) FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-fluoro-1H-indole, Beta-galactosidase, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Budiardjo, S.J, Karanicolas, J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Full and Partial Agonism of a Designed Enzyme Switch.
ACS Synth Biol, 5, 2016
|
|
5JJ1
 
 | |
5JJ3
 
 | |
8UDK
 
 | | Human Mitochondrial DNA Polymerase gamma R853A Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (24-MER), DNA (28-MER), ... | | Authors: | Park, J, Herrmann, G.K, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
8UDL
 
 | | Human Mitochondrial DNA Polymerase Gamma Binary Complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*TP*AP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*T)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Park, J, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
