3LEG
 
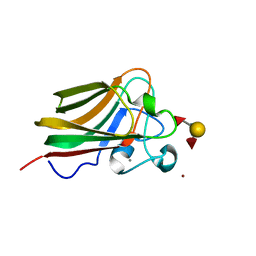 | | Lectin Domain of Lectinolysin complexed with Lewis Y Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LEK
 
 | | Lectin Domain of Lectinolysin complexed with Lewis B Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-15 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
5EZV
 
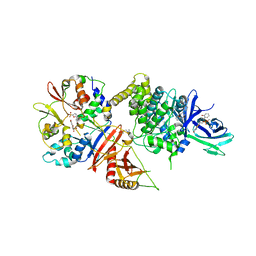 | |
6CT2
 
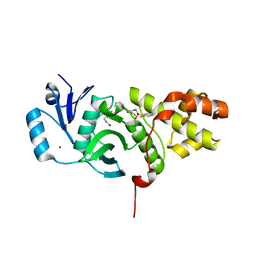 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
3DLW
 
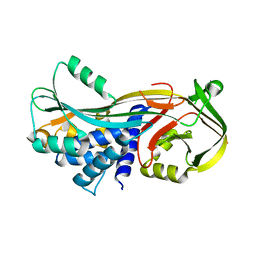 | | Antichymotrypsin | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Feil, S.C. | | Deposit date: | 2008-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and characterization of a misfolded monomeric serpin formed at physiological temperature
J.Mol.Biol., 403, 2010
|
|
6DFK
 
 | | Crystal structure of the 11S subunit of the Plasmodium falciparum proteasome, PA28 | | Descriptor: | SULFATE ION, Subunit of proteaseome activator complex,putative | | Authors: | Xie, S.C, Metcalfe, R.D, Gillett, D.L, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
3EIN
 
 | | Delta class GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1-1 | | Authors: | Feil, S.C. | | Deposit date: | 2008-09-17 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.126 Å) | | Cite: | Probing insect detoxification systems
To be Published
|
|
1NXJ
 
 | | Structure of Rv3853 from Mycobacterium tuberculosis | | Descriptor: | GLYOXYLIC ACID, L(+)-TARTARIC ACID, Probable S-adenosylmethionine:2-demethylmenaquinone methyltransferase | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-10 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Putative Methyltransferase from Mycobacterium tuberculosis: Misannotation of a Genome Clarified by Protein Structural Analysis
J.Bacteriol., 185, 2003
|
|
4Y1D
 
 | |
3SL5
 
 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6VKA
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | 2,2'-{ethane-1,2-diylbis[oxyethane-2,1-diylcarbamoyl-4,1-phenyleneethyne-2,1-diyl(5-methyl-1-benzofuran-2,3-diyl)]}diacetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | HIV Integrase core domain (IN) in complex with dimeric spanning inhibitor
To Be Published
|
|
6VLH
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-01-24 | | Release date: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | HIV Integrase core domain (IN) in complex with dimeric spanning inhibitor
To Be Published
|
|
7L23
 
 | |
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2PHM
 
 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
3PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[I104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Xiao, B. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
1GNE
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
1AQW
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1AQX
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH MEISENHEIMER COMPLEX | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
3G4O
 
 | |
3G4N
 
 | |
1PHZ
 
 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
1PGT
 
 | |
4GWI
 
 | |
