3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CWF
 
 | | Crystal structure of PAS domain of two-component sensor histidine kinase | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase synthesis sensor protein phoR | | Authors: | Chang, C, Tesar, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracytoplasmic PAS-like domains are common in signal transduction proteins.
J.Bacteriol., 192, 2010
|
|
1S7H
 
 | | Structural Genomics, 2.2A crystal structure of protein YKOF from Bacillus subtilis | | Descriptor: | ykoF | | Authors: | Zhang, R, Lezondra, L, Moy, S, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2A crystal structure of protein YKOF from Bacillus subtilis
To be Published
|
|
1SED
 
 | | Crystal Structure of Protein of Unknown Function YhaL from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Hypothetical protein yhaI, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Hypothetical Protein YhaI, APC1180 from Bacillus subtilis
To be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
3CED
 
 | | Crystal structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus | | Descriptor: | 1,4-BUTANEDIOL, Methionine import ATP-binding protein metN 2 | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus.
TO BE PUBLISHED
|
|
1SQU
 
 | |
3D0F
 
 | | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Penicillin-binding 1 transmembrane protein MrcA | | Authors: | Cuff, M.E, Mulligan, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the BIG_1156.2 domain of putative penicillin-binding protein MrcA from Nitrosomonas europaea ATCC 19718.
TO BE PUBLISHED
|
|
3LK7
 
 | | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylmuramoylalanine--D-glutamate ligase | | Authors: | Stein, A.J, Sather, A, Shakelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A
To be Published
|
|
3D1L
 
 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | Descriptor: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | Authors: | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
3D1P
 
 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
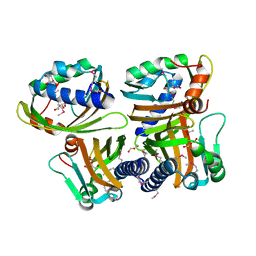
1S5A
 
 | |
1RZ3
 
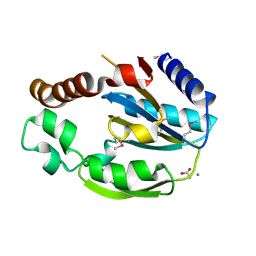 | |
3D7N
 
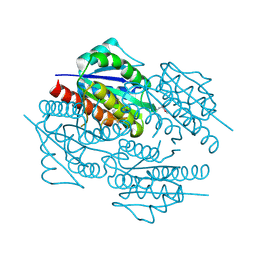 | | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens | | Descriptor: | Flavodoxin, WrbA-like protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens
To be Published, 2008
|
|
1SFS
 
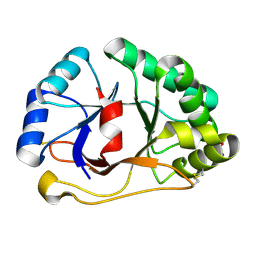 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1RXQ
 
 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
1S5U
 
 | | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Protein ybgC, SULFATE ION | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli
To be Published
|
|
1SH8
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA5026 from Pseudomonas aeruginosa, Probable Thioesterase | | Descriptor: | hypothetical protein PA5026 | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA5026 from Pseudomonas aeruginosa
To be Published
|
|
1RYL
 
 | | The Crystal Structure of a Protein of Unknown Function YfbM from Escherichia coli | | Descriptor: | Hypothetical protein yfbM | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a hypothetical protein yfbM from E. coli
To be Published
|
|
1S7I
 
 | | 1.8 A Crystal Structure of a Protein of Unknown Function PA1349 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1349 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8A crystal structure of a hypothetical protein PA1349 from Pseudomonas aeruginosa
To be Published
|
|
1INL
 
 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
