4L4T
 
 | | Structure of human MAIT TCR in complex with human MR1-6-FP | | Descriptor: | 2-amino-4-oxo-3,4-dihydropteridine-6-carbaldehyde, Beta-2-microglobulin, MAIT T-cell receptor alpha chain, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
4L4V
 
 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
7T49
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T40
 
 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4A
 
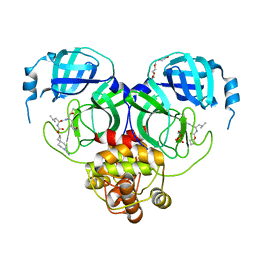 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 11c | | Descriptor: | (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
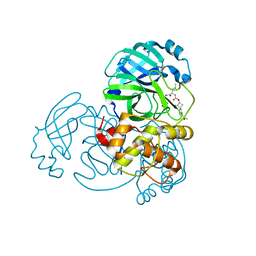 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
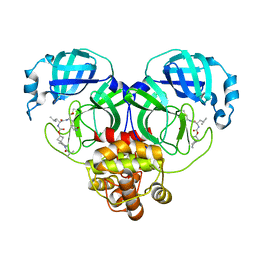 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T48
 
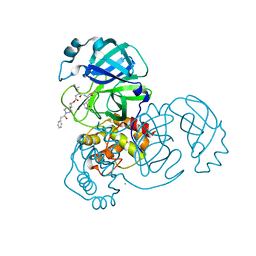 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
5FEJ
 
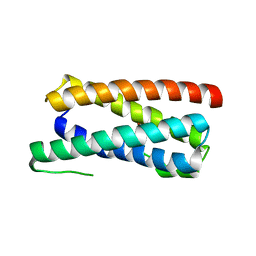 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFA
 
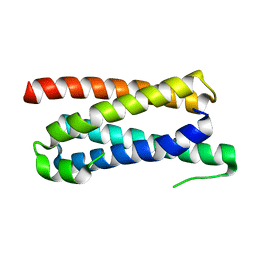 | |
7TQ7
 
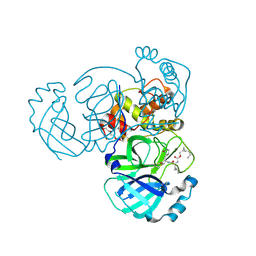 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
5FFC
 
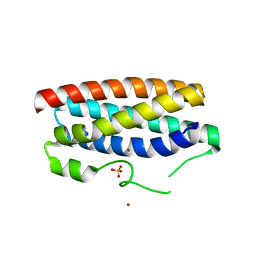 | | CopM in the Cu(II)-bound form | | Descriptor: | COPPER (II) ION, CopM, SULFATE ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFE
 
 | | CopM in the Ag-bound form (by soaking) | | Descriptor: | CopM, SILVER ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFD
 
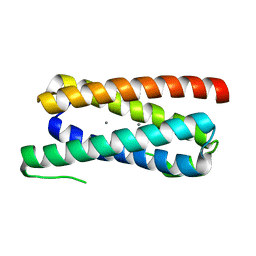 | |
7UDW
 
 | | Designed pentameric proton channel QQLL | | Descriptor: | De novo designed pentameric proton channel QQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDV
 
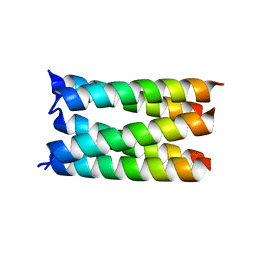 | |
7UDZ
 
 | | Designed pentameric proton channel LQLL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDY
 
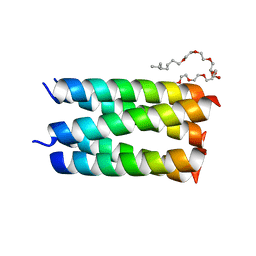 | | Designed pentameric channel QLLL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDX
 
 | | Designed pentameric proton channel QLQL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel QLQL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
5FFB
 
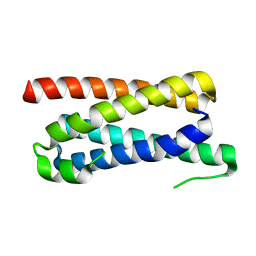 | | CopM in the apo form | | Descriptor: | CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FW5
 
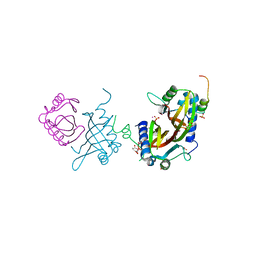 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
7DTW
 
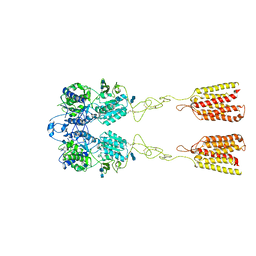 | | Human Calcium-Sensing Receptor in the inactive close-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
