2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2M72
 
 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
2M4N
 
 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
2O16
 
 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
2P5Z
 
 | | The E. coli c3393 protein is a component of the type VI secretion system and exhibits structural similarity to T4 bacteriophage tail proteins gp27 and gp5 | | Descriptor: | Type VI secretion system component | | Authors: | Ramagopal, U.A, Bonanno, J.B, Sridhar, V, Lau, C, Toro, R, Gheyi, T, Maletic, M, Freeman, J.C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2M71
 
 | | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni | | Descriptor: | Translation initiation factor IF-3 | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni
To be Published
|
|
4J3Z
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1 | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1
To be Published
|
|
4IX1
 
 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | Descriptor: | PHOSPHATE ION, hypothetical protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
4JXR
 
 | | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CITRATE ANION, ... | | Authors: | Majorek, K.A, Cooper, D.R, Porebski, P.J, Konina, K, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA
To be Published
|
|
4JWV
 
 | | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Short chain enoyl-CoA hydratase | | Authors: | Cooper, D.R, Mikolajczak, K, Cymborowski, M, Grabowski, M, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JYJ
 
 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4K9C
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID | | Descriptor: | 4-methyl-3,4-dihydro-2H-1,4-benzoxazine-7-carboxylic acid, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE and 4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-CARBOXYLIC ACID
To be Published
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4K8P
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol | | Descriptor: | (2-ETHYLPHENYL)METHANOL, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol
To be Published
|
|
4LBX
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine
To be Published
|
|
4LBG
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine
To be Published
|
|
4LC4
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with guanosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, GUANOSINE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with guanosine
To be Published
|
|
4JXU
 
 | | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP | | Descriptor: | Putative aminotransferase | | Authors: | Cooper, D.R, Cymborowski, M.T, Majorek, K.A, Niedzialkowska, E, Porebski, P.J, Stead, M, Hammonds, J, Seidel, R, Ahmed, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP
To be Published
|
|
4LNA
 
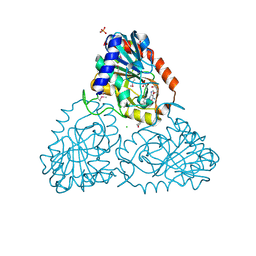 | | CRYSTAL STRUCTURE OF purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENINE, CHLORIDE ION, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362
To be Published
|
|
4LN1
 
 | | CRYSTAL STRUCTURE OF L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452 | | Descriptor: | CALCIUM ION, L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452
To be Published
|
|
4LNH
 
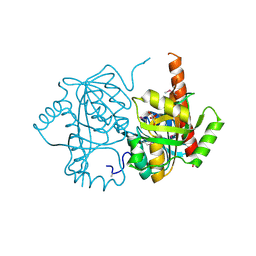 | | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520. | | Descriptor: | GLYCEROL, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520.
To be Published
|
|
4LMR
 
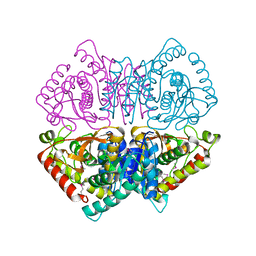 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | Descriptor: | L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
4LCA
 
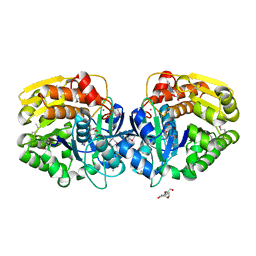 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine
To be Published
|
|
4LKR
 
 | | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-08 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437
To be Published
|
|
4JVT
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, Enoyl-CoA hydratase | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA
To be Published
|
|
