1F3S
 
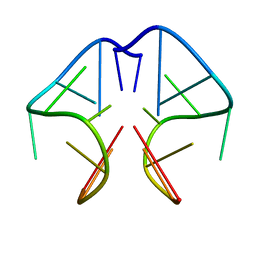 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
3QZT
 
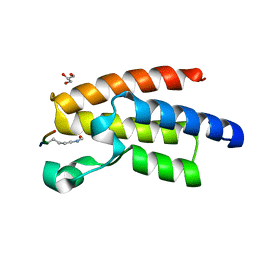 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
 | |
3RB3
 
 | |
3RBD
 
 | |
3OWW
 
 | |
3RIC
 
 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3OXB
 
 | |
3OXM
 
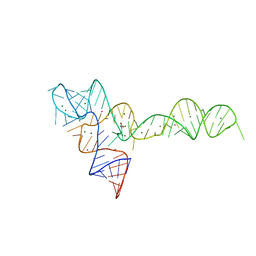 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3RIU
 
 | |
1G70
 
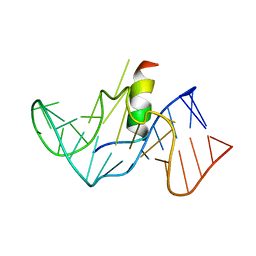 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
3RB4
 
 | |
3O7X
 
 | |
3O33
 
 | |
3NNH
 
 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3O34
 
 | |
3RB6
 
 | |
3RB0
 
 | |
3PTA
 
 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3RBE
 
 | |
3RAQ
 
 | |
1YKQ
 
 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BNV
 
 | |
