3SUG
 
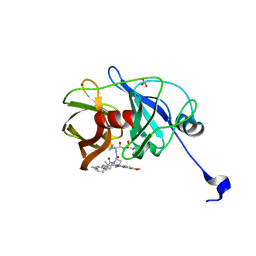 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3T3P
 
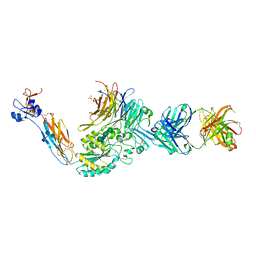 | | A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design of a High-Affinity Platelet Integrin alphaIIbbeta3 Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS
Sci Transl Med, 4, 2012
|
|
3SV9
 
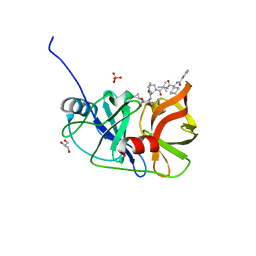 | | Crystal structure of NS3/4A protease variant A156T in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUE
 
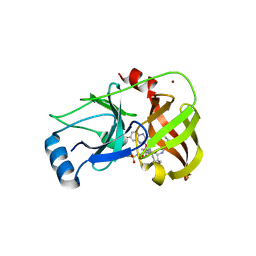 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUF
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV7
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUD
 
 | | Crystal structure of NS3/4A protease in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU2
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV6
 
 | | Crystal structure of NS3/4A protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU4
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease,NS4A protein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV8
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3T3M
 
 | | A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design of a High-Affinity Platelet Integrin alphaIIbbeta3 Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS
Sci Transl Med, 4, 2012
|
|
3SU6
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU1
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
2MAB
 
 | |
6BFA
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1553 | | Descriptor: | 1-{4-amino-3-[6-(cyclopropyloxy)naphthalen-2-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
6BE1
 
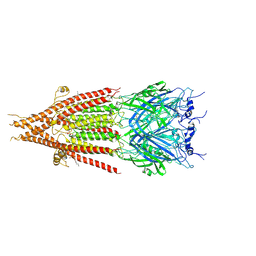 | | Cryo-EM structure of serotonin receptor | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2017-10-24 | | Release date: | 2018-02-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Cryo-EM structure of 5-HT
Nat Commun, 9, 2018
|
|
6CML
 
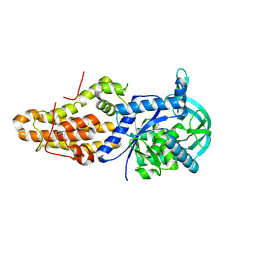 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 2093) | | Descriptor: | 1-[(5-chloro-1H-imidazo[4,5-b]pyridin-2-yl)methyl]-4-[(2-chloro-4-methoxyphenyl)methyl]-3-ethyl-1,3-dihydro-2H-imidazol-2-one, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided discovery of selective methionyl-tRNA synthetase inhibitors with potent activity against Trypanosoma brucei
Rsc Med Chem, 11, 2020
|
|
6CVX
 
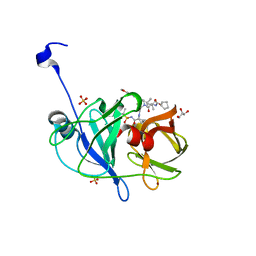 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-50 (MK-5172 linear analogue) | | Descriptor: | GLYCEROL, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-{[7-methoxy-3-(propan-2-yl)quinoxalin-2-yl]oxy}-L-prolinamide, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVY
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-21 (MK-5172 linear analogue) | | Descriptor: | 3-methyl-N-[(pentyloxy)carbonyl]-L-valyl-(4R)-4-[(3-chloro-7-methoxyquinoxalin-2-yl)oxy]-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-L-prolinamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
6CVW
 
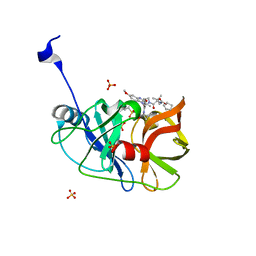 | | Crystal structure of HCV NS3/4A WT protease in complex with AJ-52 (MK-5172 linear analogue) | | Descriptor: | N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-2-ethenyl-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-4-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-L-prolinamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quinoxaline-Based Linear HCV NS3/4A Protease Inhibitors Exhibit Potent Activity against Drug Resistant Variants.
ACS Med Chem Lett, 9, 2018
|
|
2QFY
 
 | |
2QFV
 
 | |
2QFX
 
 | |
7CMO
 
 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
