9NC4
 
 | | [2,8,9-2] Shifted tensegrity triangle with an (arm,center,arm) distribution of (2,8,9) base pairs and 2 nt sticky ends | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*CP*TP*CP*TP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*GP*GP*AP*CP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-14 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (4.29 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9O8I
 
 | | [3,8,8,P-2] Shifted tensegrity triangle with an (arm, center, arm) distribution of (3, 8, 8) base pairs, 2 nt sticky ends, and 5' phosphates | | Descriptor: | DNA (5'-D(P*AP*CP*GP*GP*AP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*CP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Abi Rizk, J, Vecchioni, S, Horvath, A, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9ND2
 
 | | [1,8,10-2] Shifted tensegrity triangle with an (arm,center,arm) distribution of (1,8,10) base pairs and 2 nt sticky ends | | Descriptor: | DNA (5'-D(*AP*GP*TP*G*CP*AP*CP*TP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*GP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*GP*AP*CP*AP*GP*AP*GP*TP*GP*CP*A)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-17 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (6.61 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9NE3
 
 | | [1,8,10,P-2] Shifted tensegrity triangle with an (arm,center,arm) distribution of (1,8,10) base pairs, 1 nt sticky ends, and 5' phosphates | | Descriptor: | DNA (5'-D(P*AP*CP*GP*GP*AP*CP*AP*GP*AP*GP*TP*GP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*CP*GP*T)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-19 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (6.89 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9LK4
 
 | | Cryo-EM structure of Lhcb4.1-C2S2 PSII-LHCII supercomplex from Arabidopsis thaliana | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhou, Q, Caferri, R, Shan, J.Y, Amelii, A, Bassi, R, Liu, Z.F. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A stress-induced paralog of Lhcb4 controls the photosystem II functional architecture in Arabidopsis thaliana
To Be Published
|
|
9UEQ
 
 | | Cryo-EM structure of CT-BCCP domain of Pyruvate carboxylase from Mycobacterium tuberculosis | | Descriptor: | BIOTIN, OXALATE ION, Pyruvate carboxylase, ... | | Authors: | Singh, A, Sharma, D, Raza, M, Singh, S, Das, U. | | Deposit date: | 2025-04-09 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP
To Be Published
|
|
9O8K
 
 | | [6,8,5] Shifted tensegrity triangle with an (arm, center, arm) distribution of (6, 8, 5) base pairs, 2 nt sticky ends | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*GP*GP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Abi Rizk, J, Vecchioni, S, Horvath, A, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (6.05 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9QI6
 
 | |
9R5Y
 
 | | Crystal structure of the C-terminal domain of human TNC in complex with Adhiron 52 | | Descriptor: | Adhiron, Tenascin | | Authors: | Gaule, T.G, Trinh, C, Simmons, K.J, Tomlinson, D.C, Maqbool, A. | | Deposit date: | 2025-05-11 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Targeting Tenascin-C-Toll-like Receptor 4 signalling with Adhiron-derived small molecules - a viable strategy for reducing fibrosis in Systemic Sclerosis
To Be Published
|
|
6U57
 
 | |
9NBZ
 
 | | [5,7,7-2] Shifted tensegrity triangle with an (arm,center,arm) distribution of (5,7,7) base pairs and 2 nt sticky ends | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*CP*TP*GP*TP*GP*GP*TP*GP*C)-3'), DNA (5'-D(AP*CP*GP*GP*AP*CP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-14 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9NBV
 
 | | [5,7,8-1] Shifted tensegrity triangle with an (arm,center,arm) distribution of (5,7,8) base pairs and 1 nt sticky ends | | Descriptor: | DNA (5'-D(*TP*CP*TP*GP*AP*CP*TP*GP*TP*GP*GP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*GP*CP*AP*CP*CP*TP*GP*TP*A)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-14 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (5.97 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9U46
 
 | | The X-ray structure of N-terminal catalytic domain of Thermoplasma acidophilum tRNA methyltransferase Trm56 (Ta0931) in complex with 5'-Methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, tRNA (cytidine(56)-2'-O)-methyltransferase | | Authors: | Fukumoto, S, Hasegawa, T, Ototake, M, Moriguchi, S, Namba, M, Yamagamai, R, Kawamura, T, Hirata, A, Hori, H. | | Deposit date: | 2025-03-19 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | The X-ray structure of N-terminal catalytic domain of Thermoplasma acidophilum tRNA methyltransferase Trm56 (Ta0931) in complex with S-adenosyl-L-methionine
To Be Published
|
|
9NDH
 
 | | [4,8,8,P-1] Shifted tensegrity triangle with an (arm,center,arm) distribution of (4,8,8) base pairs, 1 nt sticky ends, and 5' phosphates | | Descriptor: | DNA (5'-D(P*AP*CP*GP*GP*AP*CP*AP*CP*GP*TP*CP*A)-3'), DNA (5'-D(P*C*TP*GP*AP*CP*GP*TP*GP*TP*GP*CP*TP*C)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-18 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (5.78 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9NWB
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-280-5I | | Descriptor: | (1R,2S,5S)-N-{(1S,2R)-1-hydroxy-1-(5-iodo-1,3-benzothiazol-2-yl)-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Hattori, S, Li, M, Das, D, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2025-03-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
9NKA
 
 | | [-1,8,12-->14] Shifted tensegrity triangle with an (arm,center,arm) distribution of (-1,8,12) base pairs and 2 nt sticky ends containing a semi-junction that becomes left-handed | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*CP*AP*CP*TP*GP*CP*TP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*AP*CP*CP*CP*GP*T)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-28 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (4.88 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
9NWA
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-277-5Cl | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-chloro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Kuwata, N, Hayashi, H, Aoki, H, Das, D, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2025-03-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
9UBG
 
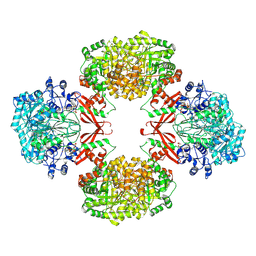 | | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Singh, A, Sharma, D, Raza, M, Singh, S, Das, U. | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP
To Be Published
|
|
8WJ2
 
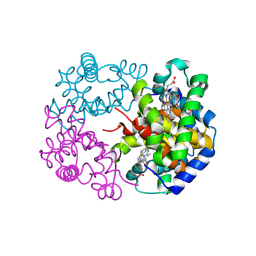 | | cryo-EM structure of human haemoglobin in deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
9OPB
 
 | | Herpes simplex virus type 1 (HSV-1) D-capsid pUL6 portal protein turrets, decamer | | Descriptor: | Capsid portal protein | | Authors: | Crofut, E.H, Kashyap, S, Stevens, A, Jih, J, Liu, Y.-T, Zhou, Z.H. | | Deposit date: | 2025-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a new capsid form and comparison with A-, B- and C-capsids clarify herpesvirus assembly
To Be Published
|
|
9O0Y
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[(2,3-dichlorophenyl)methyl]-3,5-difluoro-N-[(pyridin-3-yl)methyl]benzamide, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9MC1
 
 | | Trans-acting enoylreductase PhiaB involved in the phialotideA biosynthesis pathway | | Descriptor: | Trans-acting enoylreductase | | Authors: | Takekawa, Y, Takino, J, Sato, S, Yabuno, N, Oikawa, H, Ose, T, Minami, A. | | Deposit date: | 2025-03-17 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chain-length preference of trans-acting enoylreductases involved in the biosynthesis of fungal polyhydroxy polyketides.
Biochem.Biophys.Res.Commun., 761, 2025
|
|
6WPS
 
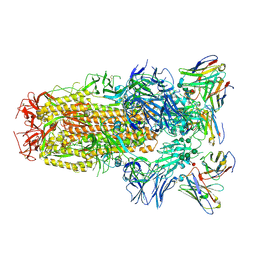 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
9NJX
 
 | | [0,7,12-->14] Shifted tensegrity triangle with an (arm,center,arm) distribution of (0,7,12) base pairs and 2 nt sticky ends containing a semi-junction that becomes left-handed | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*GP*CP*GP*TP*AP*GP*CP*TP*GP*T)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*A)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Horvath, A, Vecchioni, S, Woloszyn, K, Ohayon, Y.P, Sha, R. | | Deposit date: | 2025-02-28 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Shifted tensegrity triangles
To Be Published
|
|
